Abstract
Kaiso is a member of the BTB/POZ zinc finger family, which is involved in cancer progression, cell cycle control, apoptosis, and WNT signaling. Depending on promoter context, it may function as either a transcriptional repressor or activator. Previous studies found that Kaiso might be SUMOylated due to heat shock, but the biological significance of Kaiso SUMOylation is unclear. Here, we find that K42 is the only amino acid within Kaiso that is modified with SUMO. Kaiso is monoSUMOylated at lysine 42 in cell lines of kidney origin under normal physiological conditions. SUMOylated Kaiso can activate transcription from exogenous methylated promoters, wherein the deSUMOylated form of the protein kept the ability to be a repressor. Rapid Kaiso deSUMOylation occurs in response to hyperosmotic stress and is reversible upon return to an isotonic environment. DeSUMOylation occurs within minutes in HEK293 cells treated with 100 mm NaCl and relaxes in 3 h even in a salt-containing medium. Genomic editing of Kaiso by conversion of K42 into R42 (K42R) in HEK293 cells that resulted in fully deSUMOylated endogenous protein led to misregulation of genes associated with ion transport, blood pressure, and the immune response. TRIM25 was significantly repressed in two K42R HEK293 clones. By a series of rescue experiments with K42R and KO HEK293 cells, we show that TRIM25 is a direct transcriptional target for Kaiso. In the absence of Kaiso, the level of TRIM25 is insensitive to hyperosmotic stress. Extending our observations to animal models, we show that in response to a high salt diet, Kaiso knockout mice are characterized by significantly higher blood pressure increases when compared to wild-type animals. Thus, we propose a novel biological role for Kaiso in the regulation of homeostasis.
Similar content being viewed by others
Introduction
The complexity of biological information increases from genes to proteins. Approximately 30,000 genes ultimately result in translation of around 1 million different proteins. Protein diversity is achieved in part by post-translational modifications that often switch modified proteins from one function to another. SUMOylation involves the covalent linkage of small ubiquitin-like modifier (SUMO) polypeptide (about 100 aa) to the ε-amino group of lysine residues. The SUMOylation process is essential for viability and is detected in all eukaryotes [1, 2]. In humans, there are five different SUMO proteins. SUMO2 and SUMO3 are highly homologous and are able to form monoSUMOylated proteins and to multimerize, generating polymeric chains [3]. These chains may end with SUMO1, which never forms multimers due to lacking the key lysine residue for polySUMOylation [4]. SUMO4 is rapidly degraded under physiological conditions, but under stressed conditions, it can conjugate to a substrate protein [5]. SUMO5, a recently discovered SUMO variant, mediates the growth and disruption of PML nuclear bodies [6]. Conjugation of SUMO to lysine residues of proteins is similar to that of ubiquitin [7]. In spite of similarities between SUMO and ubiquitin modifying machinery, the functional consequences for modified proteins are different. Polyubiquitination causes rapid protein degradation, while SUMOylation usually does not change protein stability, but may influence various functional features of proteins, including cellular localization, stability, and interaction with cofactors and/or DNA.
DNA methylation is essential to mammalian development. It has a critical role in cell processes, such as the regulation of transcription, X inactivation, heterochromatin formation, imprinting, and tissue-specific gene expression. Methylated DNA attracts methyl-DNA-binding proteins that are implicated in the repression of transcription [8]. Kaiso (ZBTB33) is one of these methyl-DNA-binding proteins. It is involved in the methyl-dependent repression of gene transcription by recruiting corepressors N-CoR and SMRT [9, 10]. In addition to associating with methylated DNA, Kaiso can bind CTGCNA sequences, but not hydroxymethylated DNA [11,12,13]. Treatment with DNA damaging agents and hypoxia lead to increased expression of Kaiso [14, 15]. Recently, it was shown that Kaiso might also participate in transcriptional activation of proapoptotic genes by p53 in response to DNA damage. Kaiso interacts with the p53-p300 complex and influences p53 acetylation [15].
Several reports showed that Kaiso may be SUMOylated upon heat shock [16, 17]. However, nothing is known about the functional significance of Kaiso SUMOylation. Here, we show that Kaiso exists as a monoSUMOylated protein, SUMOylation occurs at Lys42 within the BTB domain, deSUMOylation switches Kaiso from a transcriptional activator to a repressor upon hyperosmotic stress.
Materials and methods
Cell cultures and transient transfections
HEK293, COS-7, Caki-1, and (Mbd2−/−, Kaiso−/y) [18] were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mm l-glutamine. HEK293 cells were transfected with Profection@ (Promega), MBD2−/−, Kaiso−/y. Cells were transfected with Lipofectamine2000 (ThermoFisherScientific) according to the manufacturer’s protocol. siRNA control (GAPDH) and Kaiso (sense 5′-GGCAGUUAUUAGGAGUGAtt antisense 5′-UCACUCCUAAUAACUGCCCtg) were transfected with Lipofectamine RNAiMAX (ThermoFisherScientific). Cells were typically harvested 48 h post-transfection for further analysis.
Methylation-dependent repression assay
The design of the assay was described previously [19]. Briefly, a luciferase reporter plasmid (pGL2-promoter, Promega) that contained 11 CpGpCpG sites upstream of SV40 promoter was methylated by methylase SssI (NEB) according to the manufacturer’s instructions. Completeness of methylation was defined by methyl-sensitive restriction enzyme HpaII digestion (ThermoFisherScientific) and followed by separation of the fragments in agarose gel. Each well of a 12-well plate with MBD2−/−, Kaiso−/y cells was transfected with 1 μg of methylated or unmethylated reporter plasmid and 25 ng of a control plasmid encoding Renilla (pRenilla, Promega). For titration with Kaiso and Kaiso K42R vectors, two concentrations, 25 and 75 ng per well, were used. SUMO1-GFP expression vector was co-transfected at a concentration of 50 ng per well. Luciferase levels were measured after 48 h using the Dual Luciferase Assay Kit according to the manufacturer’s instructions (Promega). Corrected values were obtained according to the formula: (luciferase sample − luciferase control)/(Renilla sample − Renilla control), where control values are obtained from untransfected cells. Relative luciferase values were defined as the corrected value obtained using a methylated test plasmid divided by the corrected value obtained using the unmethylated test plasmid multiplied by100 (Supplementary Table 1).
KBS-dependent repression assay
The KBS (Kaiso-binding sequence) luciferase reporter plasmid was generated by inserting four tandem copies of an oligonucleotide harboring the consensus KBS element (CTGCCA) (5′-TGTGTGCTTCCTGCCAATAACGATG-3′) between the SacI and XhoI in the multiple cloning site of the pGL2-promoter reporter vector. pGL2-KBSx4 (400 ng) was cotransfected with Renilla (10 ng) into each well of 24-well plate with HEK293 KO cells that were deficient for Kaiso. For titration with Kaiso, Kaiso K42R and SUMO1-GFP vectors, the same concentrations were used as previously described in the section “Methylation-dependent transcriptional assay”. For control we used pGL2-promoter vector without KBS sites. Corrected values were obtained according to the formula: (luciferase sample − luciferase control)/(Renilla sample − Renilla control), where control values are obtained from untransfected cells. Relative luciferase values were defined as the corrected value obtained using a pGL2-KBSx4 plasmid divided by the corrected value obtained using the pGL2-without KBS plasmid multiplied by 100 (Supplementary Table 1).
Antibodies
Following reagents were used in this study: anti-Kaiso polyclonal rabbit antibodies (ab’s) (kindly gifted by Dr. A. Reynolds), anti-SUMO1 (sc5308, Santa Cruz, sc-32058 Abcam), anti-SUMO2/3 (sc32873, Santa Cruz), anti-GFP (G1544, Sigma), anti-HA (A190-108A, Bethyl), anti-SUMO1 agarose (sc-5308 AC, Santa-Cruz), anti-HA agarose (A2095, Sigma), anti-TRIM25 (ab86365), anti-actin (ab8227), anti-PCNA (sc-25280, Santa-Cruz), anti-H3K4me3 (ab1012, Abcam), anti-H3K27ac (ab4729, Abcam), anti-H3K9me3(ab8898, Abcam).
Stress treatment
To induce stress, media was removed from cells and quickly replaced with fresh media supplemented with the indicated concentration of osmolyte (NaCl, sorbitol) or other stress agents (ethanol, H2O2). For treatment by gradient increased osmolality, cells were incubated in 20 mm HEPES pH7.9 with the corresponding concentration of NaCl. Cells were stressed during the times indicated in figures and then used for analysis. For heat shock, cells were incubated at 42 °C for 1 h and then harvested. For UV-induced stress, cells were treated with UV 254 nm for 90 s with 5 min of recovery (approximately 100,000 microjoules). Control cells were treated similarly without the addition of stress agents.
Preparation of nuclear extracts
Nuclear extracts were purified as described in ref. [20] with slight modifications. N-ethyl maleimide protease inhibitor (NEM) was added to hypotonic buffer A and buffer C to a final concentration 10 mm.
Plasmids and vectors
BTB/POZ domains of Kaiso orthologs (human, fish, frog) were cloned in pFLAG-CMV2, and an HA tag was subcloned at the 3′ end of the BTB/POZ cDNA. Expression vectors for SUMO1-GFP, pcDNA3-UBC9 were kindly provided by Dr. R. Niedenthal. Sequences of all constructs were verified by Sanger DNA sequencing. By subcloning an expression vector with a chimeric protein that contains E2–SUMO ligase Ubc9 and the N-terminal fragment of Kaiso (1–490 aa)—KaisoΔZF was generated.
Ligation of BbsI-digested pSPCas9(BB)-2A-Puro (PX459) (Addgene #48139) plasmid [21] with annealed sgRNA oligo insert: Crisp_ZBTB33_for 5′-CACCGTGTGGAAGACCGAAAATTCC, Crisp_ZBTB33_rev 5′-AAACGGAATTTTCGGTCTTCCACAC. The plasmid was verified by sequence analysis.
Immunoprecipitation
Immunoprecipitation was performed in the presence of 10 mm NEM with anti-SUMO1, anti-SUMO2/3, Kaiso, anti-SUMO1 agarose, control IgG antibodies and HA-agarose.
ChIP
Crosslinking for ChIP was performed by EZ-ChIP (17–371, Millipore) with anti-Kaiso rabbit pAb’s or IgG antibodies according to the manufacturer’s instructions. After DNA purification, 30 cycles of PCR were run with primers specific to the promoter region of human TRIM25 gene: for 5′-GACCTTCGCCAGCTCATGGG, rev 5′-ACACAACTGCTGCACCCGC.
Animal care
The experiment was performed on C57BL/6J (wild-type—wt) and C57BL/6Kaiso−/y (Kaiso-deficient—KO) mice. C57BL/6Kaiso−/y mice were obtained in Adrian Bird’s lab (Edinburgh University) by genetic knockout of the Kaiso gene in C57Bl/6J animals [22], C57 BL/6 J mice were obtained from Jackson Laboratories and transferred to the Center for Genetic Resources of Laboratory Animals at the Institute of Cytology and Genetics, Siberian Branch, RAS (RFMEFI61914X0005 and RFMEFI62114Х0010). All procedures were conducted under Russian legislation according to Good Laboratory Practice, inter-institutional bioethical committee guidelines and the European Convention for the protection of vertebrate animals used for experimental and other scientific purposes. All procedures were approved by the bioethical committee (protocol number 33 20.04.2016). All efforts were made to minimize the number of animals used and their suffering. All animals used had specific pathogen-free status. Animals were randomly allocated to groups that received either a high salt diet (HSD) (8% NaCl, n = 6) or a standard diet (SD) for 3 weeks (n = 6). All animals were housed under a 14 h/10 h light/dark photoperiod with 22–24 °C room temperature, 30–60% humidity, and 14–16 air changes per hour; SD (SSniff, Germany) or HSD and water were provided ad libitum.
A measure of blood pressure
Blood pressure was measured in 8-week-old male mice before beginning the diet and in 11-week-old mice after 3 weeks of HSD by tail-cuff manometry using a CODA-2 non-invasive blood pressure monitoring system (Kent Scientific, Torrington, CT) as described previously [23]. A measure of blood pressure was conducted between 1600 and 2000 hours (dark time period) by the same two observers each time. Mice were adapted to our method of measuring blood pressure for 2 days. Then, the blood pressure of mice was determined over a 5-day period. The mice were restrained in a plastic tube restrainer. Then, occlusion and volume-pressure recording cuffs were placed over their tails, and the mice were allowed to adapt to the restrainer for 5 min prior to the blood pressure measurement protocol. After the 5 min adaptation period, blood pressure was measured for ten acclimation cycles followed by 20 measurement cycles. Mice were warmed by heating pads during the acclimation cycles to ensure sufficient blood flow to the tail. The animals were monitored closely throughout the measurement protocol and removed from restraint as soon as possible upon completing the measurement protocol. Only datasets from mice characterized by a stable mean arterial pressure and a pulse pressure of 20 mmHg were included for further analysis. This experiment was repeated twice.
Cryosections
Cryosections were prepared as described in ref. [24]. Immunohistochemistry was made with anti-Kaiso rabbit antibodies.
CRISPR/CAS9-based gene editing
CRISPR/CAS9–based editing was performed as described in ref. [21]. Briefly, HEK293 were seeded on a 12-well plate. Cells were transfected with Lipofectamine 2000 with PX459-ZBTB33 plasmid alone to generate KO cells (frame shift) or with ssODN 5′-CGTGGCCATGGACTCTTCTGTGATGTTACCGTTATTGTGGAAGACCGACGCTTCCGCGCTCACAAGAATATTCTTTCAGCTTCTAGTACC to generate K42R cells (single nucleotide substitution). Puromycin was applied after 24 h at a concentration of 2 μg ml−1. Cells were incubated for 72 h, passaged into a 96-well plate at a density of 1 cell per 2 wells on medium without puromycin. For further study, we have chosen two clones for each experiment (frame shift that mimics KO cells, and single nucleotide substitution, that forms K42R cells). The substitutions (K42R) and frame shift (KO) were confirmed by Sanger sequencing of the corresponding amplicons obtained from PCR with genomic DNA.
RNA purification and RNA-seq
Total RNA was extracted from cells with TRIzol reagent according to the manufacturer’s instructions (Invitrogen) in triplicate. Quality was checked with BioAnalyser and RNA 6000 Nano Kit (Agilent). PolyA RNA was purified with Dynabeads® mRNA Purification Kit (Ambion). An Illumina library was made from polyA RNA with NEBNext® mRNA Library Prep Reagent Set (NEB) according to the manual. Sequencing was performed on HiSeq1500 with a 50 bp read length (BioProject ID PRJNA386575). At least 40 million reads were generated for each sample. Reads were mapped to hg19 genome with tophat2 version 2.1.1. Gene models of non-overlapping exonic fragments were taken from RefSeq database. For each gene, we found total coverage by mapped reads in each sample with bedtools coverage (version 2.26.0). Differential expression analysis was performed by applying default read count normalization (estimateSizeFactors) and performing per-gene negative binomial tests (nbinomTest), implemented in DESeq R package (version 1.22.0), with default parameters 15. Enriched Gene Ontology terms were identified using DAVID bioinformatics resources with genes pre-ranked according to their fold change induced by the presence of K42R Kaiso [25].
Quantitative RT-PCR (qRT-PCR)
Total RNA (2 μg) was treated with 2 U DNase I (ThermoFisherScientific) and reverse transcribed with random hexamers as primers and Revertaid First Strand cDNA synthesis kit (ThermoFisherScientific). qRT-PCR was performed using TaqMan probes on CFX96 (BioRad) instrument. Amplification of GAPDH transcript served as RNA controls for relative quantitation. Used primers: GAPDH (for: 5′-ACCAGGGCTGCTTTTAACTC-3′; rev:5′-AGATGGTGATGGGATTTCCA; probe: FAM 5′-TCATTGACCTCAACTACATGGTTTACA-3 RTQ1), TRIM25 (for: 5′-GCCTGGTGGAGCATAAGACC-3′; rev: 5′-TCTGACTGTACATGACAGTTAGT-3′; probe: FAM 5′-CCTGGAGGCCACCCTGAGGCAC-3′ RTQ1).
Apoptosis analysis
Cells were seeded in a six-well plate and treated with culture medium supplemented with 100 mm NaCl for 24 h. Apoptosis analysis was performed with AnnexinV-FITC kit (BioVision) according to the manufacturer’s instructions.
Results
Kaiso is SUMOylated under normal conditions
First, we checked whether Kaiso is SUMOylated under normal conditions. Western blot analysis of lysates from HEK293, Caki-1, COS-7 cell lines showed a significant change in mobility of Kaiso (Fig. 1a). The specificity of detected bands was confirmed by western blot with lysates from a Kaiso knockout HEK293 cell line (KO cells) produced by the CRISPR/CAS9 editing system (Fig. 1a). This change in mobility (10–20 kDa) was attributed to post-translational modifications. We hypothesized that the shift in mobility may be due to the covalent link between Kaiso and SUMO, since SUMO typically adds 15–17 kDa to the size of the targeted protein [26]. We compared the pattern of Kaiso modifications in nuclear extracts prepared with or without SUMO proteases inhibitor NEM. In the presence of NEM, anti-Kaiso antibodies detected the extra band in HEK293 cells (Fig. 1b, left panel). The same was observed for nuclear extracts purified from Kaiso-GFP transfected cells (Fig. 1b, right panel). To determine if this band corresponds to the SUMOylated protein, total extracts from HEK293 cells were immunoprecipitated with SUMO-1 or SUMO-2/3 antibodies with or without NEM. Only SUMO-1 antibodies pulled down Kaiso (Fig. 1c). Both SUMOylated and nonSUMOylated Kaiso were precipitated with SUMO-1 antibodies. This might be due to the intrinsic feature of the BTB/POZ domain to dimerize [27], resulting in a multimeric complex containing both SUMOylated and nonSUMOylated forms of the protein. However, we cannot exclude the possibility of non-covalent interactions of Kaiso with SUMO1. The latter is supported by Kaiso precipitation with SUMO1 without NEM (Fig. 1c). Western blot analysis with anti-SUMO-1 antibodies after precipitation with Kaiso antibodies confirmed that the slower migrating band corresponds to the SUMOylated form of Kaiso. Extracts prepared without SUMO inhibitor did not produce the slower migrating band. To confirm the role of SUMO in Kaiso modifications, HEK293 cells were co-transfected with Kaiso-GFP and SUMO-1, -2, -3 expression vectors. Immunoprecipitation with NEM and western blot analysis using anti-SUMO antibodies revealed that Kaiso may be modified by all three forms of SUMO (Fig. 1d, e). The modified form was not detected in the western blots probed with anti-SUMO antibodies in NEM-deficient lysates.
SUMOylation of Kaiso protein under normal conditions. a Cells Caki-1, HEK293, COS-7 and two Kaiso knockout clones were lysed in loading buffer and analyzed by SDS-PAGE and western blotting, and probed with anti-Kaiso antibody to detect Kaiso and its post-translationally modified forms. Kaiso (>100 kDa) is post-translationally modified under normal conditions. b Nuclear extracts were prepared with or without SUMO isopeptidase inhibitor NEM from HEK293 (left panel) and from HEK293 transfected with Kaiso-GFP (right panel) and analyzed by SDS-PAGE, western blotting, and stained with anti-Kaiso antibody. In the presence of NEM, an additional band was detected. c The whole-cell lysates were immunoprecipitated with anti-Kaiso, anti-SUMO1, anti-SUMO2/3 with or without NEM. Western blot analysis revealed that Kaiso and SUMO1 co-immunoprecipitated. Kaiso is SUMOyaled by SUMO1. d, e Kaiso-GFP was transfected into HEK293 cells alone or with SUMO1, 2, 3,-GFP plasmids. Cells were lysed in loading buffer or immunoprecipitated with Kaiso antibodies with or without NEM and analyzed by western blot. Kaiso may be monoSUMOylated by all forms of SUMO
SUMO is attached to Kaiso at K42 within BTB/POZ domain
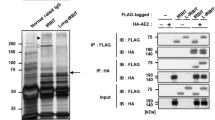
Next, we identified potential SUMOylation sites in Kaiso. Analysis of the amino acid sequence of Kaiso did not reveal any SUMOylation consensus motif, i.e. ΨKXE [28]. Primary mapping was accomplished using the UFDS system: Ubc9 SUMO conjugating enzyme Fusion-Directed SUMOylation that increases the amount of the SUMOylated form of protein independently of necessary SUMO ligase [29]. We showed that C-terminal part of Kaiso with three zinc fingers is not necessary for Kaiso modification (Fig. S1a,b). Thus, the SUMOylation site should be localized at the N-terminus of Kaiso. The evolutionarily conserved and structured domain within this N-terminal region is the BTB/POZ domain (1–123 aa). Co-transfection of Flag/HA-tagged BTB domain of Kaiso and SUMO1-GFP expression vectors produced a SUMOylated form of the domain (Fig. 2a). Therefore, SUMOylated lysine (or lysines) should be located within the BTB/POZ domain.
Kaiso is modified by SUMO-1 on 42 K in BTB/POZ domain. a HEK293 were co-transfected with Flag-BTB-HA, SUMO1-GFP, EGFP as indicated. Flag-BTB-HA from whole-cell lysates was immunoprecipitated with anti-HA agarose in the presence or absence of NEM. Immunoblotting with GFP antibodies revealed that the BTB/POZ domain is modified by SUMO-1. b, c Nuclear extracts were prepared with or without NEM from HEK293 cells transfected with Kaiso-GFP, K42R Kaiso-GFP, and SUMO1-GFP (b), and from cells with endogenous K42R form of Kaiso (c). The anti-Kaiso antibody detected an additional band in nuclear extracts prepared with NEM from HEK293 cells transfected with wild-type Kaiso, but not with the K42R mutant form. d Immunoprecipitation with anti-Kaiso antibody from clones with endogenous K42R Kaiso
The functional properties of the BTB/POZ domain are evolutionarily conserved [30, 31]. Hypothetically, lysines that are subjected to SUMOylation must be conserved between different organisms as well. To test this, BTB/POZ domains of Kaiso orthologs from Xenopus Laevis and Danio Rerio were coexpressed with SUMO1-GFP. Both BTB/POZ domains were SUMOylated (Fig. S1c). The most strongly conserved lysines within BTB/POZ domains among four species (human, mouse, frog, fish) were selected, i.e. K5, K42, K124 (Fig. S1d). Then, we substituted K5, K42, and K124 for arginines R5, R42, and R124 correspondingly. Mutated BTB/POZ domains with Flag/HA tags were coexpressed with SUMO1-GFP and western blotted with GFP antibodies (Fig. S1e). SUMOylated BTB/POZ domain was absent only in the K42R mutant. To prove that K42 is the major SUMOylation site, the same point mutation was introduced into the full-length human Kaiso-GFP expression vector. In contrast to wild-type form, the K42R mutant form of Kaiso did not produce any SUMOylated bands, regardless of the presence or absence of NEM (see Fig. 2b). Moreover, even when exogenous SUMO1 was coexpressed with the K42R mutant, the modification was not detected (Fig. 2b). Using the CRISPR/CAS9 system we generated HEK293 cell line with biallelic point mutation K42R in endogenous Kaiso locus. Western blot analysis showed a loss of endogenous Kaiso SUMOylation in nuclear extracts prepared from K42R cells, although we still observed a very weak slower migrating band (Fig. 2c). Immunoprecipitation with Kaiso antibodies confirmed that mutant K42R form cannot be SUMOylated (Fig. 2d).
Kaiso K42R retains ability to repress transcription
Kaiso has dual DNA-binding specificity: it may bind either methylated motifs that contain mCpG or unmethylated DNA with core sequence CTGCNA [11]. In order to determine whether SUMOylation/deSUMOyaltion of Kaiso has any effect on its transcriptional properties, we performed two sets of transient transfection experiments.
First, we co-transfected Kaiso or its mutant form K42R along with methylated or non-methylated reporter genes into (Mbd2−/−, Kaiso−/y) mouse fibroblasts. We used this cell line to avoid repression of the methylated reporter due to activity of endogenous methyl-DNA-binding proteins and to minimize the effect of endogenous Kaiso [32]. Expression of exogenous intact Kaiso and K42R mutant reduced transcription to 40 and 20% respectively at the highest used concentration (Fig. 3a, left panel). Thus, K42R mutation did not affect repression ability of the protein. Cotransfection of a SUMO-1 expression vector switched Kaiso into a mild transcriptional activator (150%), and had little influence on the repressive properties of K42R (30%) (Fig. 3a, right panel).
Kaiso K42R retains ability to repress transcription in a transient transfection assay. Kaiso expression constructs were co-transfected with Sss1 methylated or nonmethylated SV40-luciferase reporter into mouse cells (Mbd2−/−, Kaiso−/y) (a) or cotransfected with KBS-pGL2-PV in HEK293 KO cells (b). Relative reporter activity is presented as a percentage (methylated reporter expression/nonmethylated reporter expression×100) (a) or is presented as a KBS reporter expression/pgl2withoutKBS reporter expression×100. Results are the average of at least three experiments. Mean ± standard error of mean (n = 3, Student’s t test)
Second, we evaluated the effect of SUMOylation on a functional switch of Kaiso transcriptional properties in a methylation independent system by determining transcriptional activity using KBS luciferase reporter plasmid in HEK293 KO cells. Intact Kaiso as well as K42R mutant represses Luciferase reporter (Fig. 3b, left panel). However, as soon as exogenous SUMO is expressed intact Kaiso starts to activate the reporter transcription whereas K42R induced repression is almost insensitive to the presence of SUMO (Fig. 3b, right panel).
We concluded that overexpression of SUMO may switch Kaiso from repressor to activator. At the same time K42R mutant functions as repressor independently of presence of exogenous SUMO. This property of Kaiso works equally well when tested in assays that rely on its different DNA binding specificity—KBS and mCpG’s.
Osmotic stress induces Kaiso deSUMOylation
Since SUMOylation/deSUMOylation is a rapidly changing modification under different stress conditions, we studied Kaiso SUMOylation in various stress-induced systems. We exposed HEK293 cells to heat shock, hyperosmotic stress, hydrogen peroxide, and ethanol. Western blot analysis of total cell lysates with anti-Kaiso antibodies revealed significant differences between the SUMOylated and deSUMOylated fractions of the protein upon hyperosmotic stress (Fig. 4a), while it had no effect on the K42R form of Kaiso (Fig. S2a). The same deSUMOylation was detected in Caki-1 and COS-7 cell lines (Fig. S2b). Thus, we focused on the effect of NaCl-induced osmotic stress on post-translational modification of Kaiso.
Osmotic stress induces changes in Kaiso modification. Kaiso is deSUMOyalted upon NaCl treatment. a HEK293 cells were exposed for 5 min to ethanol, H2O2, 400 mm NaCl, urea, or to temperatures of 42 °C for 1 h. b HEK293 and K42R cells were exposed to 400 mm NaCl, 0.4 M sorbitol for 15 min. After treatment with NaCl, cells were recovered for 15 min in DMEM/FBS. c HEK293 cells in DMEM/FBS were incubated with NaCl (100 mm) during indicated time intervals. Western blot analysis showed the percentage of modified forms of Kaiso after 100 mm NaCl treatment (n = 3, mean ± standard error of mean, pvalue indicated on graph). a–c Cells were lysed in loading buffer or immunoprecipitated with anti-Kaiso antibody, and analyzed by SDS-PAGE and western blotting. d Blood pressure measurements in wild-type and Kaiso knockout mice after 3 weeks on a high salt diet (n = 8, mean ± standard error of mean, ***pvalue < 0.0001, **pvalue < 0,01, Student’s t test)
The effect of hyperosmotic stress on Kaiso modifications was examined by gradiently increased osmolality. We observed major changes in Kaiso modification from 175 to 400 mm NaCl-induced stress (Fig. S2c) when the cells were treated for 5 min. To exclude the possibility that modification of Kaiso is sensitive to increased expression of alpha-Na+-K+-ATPase [33], the cells were treated for 15 min with media that was supplemented with 0.4 m sorbitol, which works as an alternative chemically distinct osmolyte. The observed effect with sorbitol was the same as the effect with 0.4 m NaCl (Fig. 4b). Thus, the effect is caused by hypertonicity. Returning the cells to normal conditions for 15 min after hyperosmotic stress led to the recovery of Kaiso modification (Fig. 4b). K42R cells did not produce any changes when their extracts were probed with Kaiso and SUMO-1 antibodies. The time-course of NaCl treatment suggested that at 100 mm NaCl, cells quickly deSUMOylate Kaiso (in 5 min), but then SUMOylation is restored over a period of 3 h to almost 100% of initial SUMOylation levels (Fig. 4c). The time-course of sorbitol treatment confirmed the same effects that were observed for NaCl (Fig. S2d). Thus, Kaiso is rapidly deSUMOylated upon salt treatment, and depending on salt concentration, SUMOylation of Kaiso may recover (at low salt Fig. 4c) or be lost (at high salt Fig. S2d).
Salt-sensitive hypertension in Kaiso knockout mice
Previously, Kaiso expression was detected in many tissues and cell types, predominantly in epithelial cells that pave organ borders with environmental fluids (intestines, cornea, skin, bladder, etc.) [24]. In turn, we have shown that Kaiso is expressed also in subsets of kidney cells that resemble distal tubules (Fig. S3a). Therefore, the pattern of Kaiso expression implies the possibility of its participation in the response to hyperosmotic stress.
To confirm the biological importance of hyperosmotic stress in vivo on the function of Kaiso, we fed wild-type and Kaiso knockout mice a high salt diet. The blood pressure of wild-type and Kaiso-deficient mice was equal under a normal diet (Fig. 4d). However, when the animals were switched to a high salt diet (8% of salt in the ration)) for 3 weeks, blood pressure showed a dramatic increase in Kaiso knockout mice compared to wild-type mice (Fig. 4d). Thus, the absence of Kaiso predisposed mice to increased blood pressure upon a high salt diet.
DeSUMOylated form of Kaiso represses TRIM25 transcription
Finally, we explored the functional role of Kaiso deSUMOylation on gene transcription by measuring the digital amount of mRNA transcripts. We found 558 downregulated (pvalue < 0.001, twofold) and 494 upregulated (pvalue < 0.001, twofold) genes in K42R (clone1) compared to HEK293 cells (Supplementary Table 2). GO (Gene Ontology) term analysis of downregulated mRNA in the K42R mutant cell line identified transcripts associated with the inflammatory response, ion transport, and regulation of blood pressure (Supplementary Table 3). GO analysis of upregulated mRNA revealed genes associated with protein folding, RNA processing, and mitochondrial organization (Supplementary Table 3). Among the top repressed genes in the K42R mutant cell line, we found TRIM25. TRIM25 was first discovered as an estrogen-responsive finger protein [34]. It has been implicated in the proliferation of many cancer cell types [35]. Additionally, it was shown that TRIM25 may escape from the estrogen-mediated control of expression [36].
Downregulation of TRIM25 in both K42R clones was confirmed by real-time PCR and western blot (Fig. 5a, b). We observed stronger TRIM25 repression in K42R clone1 than in clone2. Deficiency of Kaiso led to TRIM25 upregulation (Fig. 5a, b). We next tested whether Kaiso occupies the TRIM25 promoter. ChIP analysis showed that Kaiso binds the TRIM25 promoter region in both HEK293 and K42R cell lines (Fig. 5c). Therefore, the nonSUMOyalted form of Kaiso can repress TRIM25 transcription in cellular models.
Kaiso regulates TRIM25 expression. a qPCR-RT analysis of TRIM 25 expression in HEK293, K42R and Kaiso-deficient clones with Taqman probe. HEK293. b Clones with a mutation on 42 K in Kaiso and Kaiso knockout were lysed in loading buffer or immunoprecipitated with anti-Kaiso antibody, and analyzed by western blotting. TRIM25 is downregulated in K42R clones. Cells were treated with 250 mm NaCl 3 h, then lysed in loading buffer and analyzed by western blotting (d, e) and qPCR-RT (a). TRIM25 is downregulated in HEK293 cells upon hyperosmotic stress, whereas a deficiency of Kaiso prevents TRIM25 downregulation upon hypertonicity. c Kaiso binds within the TRIM25 promoter region. PCR after ChIP with anti-Kaiso antibody on the promoter region of TRIM25
To study how changes in Kaiso deSUMOylation influence TRIM25 transcription, we performed hyperosmotic treatment of HEK293, K42R, and KO cells (Fig. 5). A significant decrease of TRIM25 expression was detected in HEK293 cells, along with Kaiso deSUMOylation. The level of TRIM25 was unaltered in K42R mutant cell lines during treatment (Fig. 5a, d). Kaiso deficiency effectively abolished salt-induced suppression of TRIM25 mRNA (Fig. 5a, e). These results indicate that hyperosmotic stress causes suppression of TRIM25 transcription through deSUMOylated Kaiso.
To show the specificity of Kaiso to the modulation of TRIM25 activity, we performed a series of rescue experiments. First, we expressed exogenous Kaiso-GFP in KO cells. Restoring Kaiso level resulted in a decrease of TRIM25 transcription (Fig. 6a). Second, we used siRNA specific to Kaiso to deplete its mRNA in K42R cells. In spite of downregulated Kaiso, we did not observe the expected increase in TRIM25 transcription (Fig. 6b). We hypothesized that K42R Kaiso established such an unfavorable chromatin state with the TRIM25 promoter that even when Kaiso K42R level was significantly decreased by siRNA, the promoter remained in an inactive state due to epigenetic factors. ChIP analysis demonstrated that the TRIM25 promoter in HEK293 and KO cells is enriched with active histone marks (H3K4me3, H3K27ac) and depleted of inactive marks (H3K9me3). K42R cells were characterized by a loss of active histone marks within the TRIM25 promoter that were replaced by repressive H3K9me3. siRNA knockdown of Kaiso in K42R cells did not erase the H3K9me3 mark, suggesting that deSUMOylated Kaiso may participate in the establishment of inactive chromatin around the TRIM25 gene, but not in the maintenance of this chromatin state (Fig. 6c).
Effect of Kaiso deSUMOylation on chromatin marks and apoptosis. Effect of Kaiso-GFP overexpression in Kaiso-deficient cells (a) and Kaiso knockdown in K42R mutant cells (b) on TRIM25 expression. c ChIP analysis of chromatin marks (H3K4me3, H3K27ac, H3K9me3) on TRIM25 promoter in cells with different Kaiso status (wild type—HEK293, Kaiso deficient—Kaiso KO clone1, non-SUMOylated Kaiso—K42R clone1, Kaiso recovery in Kaiso KO clone1, Kaiso knockdown in K42R clone1). Active marks H3K4me3 and H3K27ac are lost in cells with nonSUMOylated Kaiso and are not restored upon knockdown of Kaiso. d Data analysis of AnnexinV-FITC, PI staining of cells under normal conditions and after incubation with 100 mm NaCl during 24 h (n = 3, mean ± standard error of mean, Student’s t test). e Schematic model of Kaiso action on promoter activity in dependence from SUMOylation
Kaiso K42R mutation enhances osmos-induced apoptosis in HEK293 cells
Downregulation of TRIM25 results in increased apoptosis of lung cancer cells treated with doxorubicin [37, 38]. We hypothesized that the level of osmos-induced apoptosis will be different in K42R cells compared to HEK293 and KO cells, depending on the amount of TRIM25 mRNA in each cell type. To test this, we stressed cells with 100 mm NaCl for 24 h. This treatment led to rapid but transient Kaiso deSUMOylation (see about SUMOylation recovery above and Fig. 4c). Cells staining for annexinV after hyperosmotic treatment showed that HEK293 cells exhibited lower levels of apoptosis (14.7%) compared to K42R clones: clone1 = 21% and clone2 = 32% (Fig. 6d, Fig. S4). Kaiso-deficient cells had a level of apoptosis almost identical to that of HEK293 cells. When KO cells were rescued with expression of exogenous intact Kaiso, we detected higher levels of apoptosis that were in line with downregulation of TRIM25 (compare panels a and d in Fig. 6, Fig. S5). Kaiso knockdown by siRNA in K42R cells did not change the level of apoptosis, which agrees with the lack of change in TRIM25 transcription after treating K42R cells with siRNA. Thus, we confirm that a dominant negative Kaiso mutant, which is unable to be SUMOylated, downregulates TRIM25 mRNA levels and predisposes the cells to higher apoptosis upon hyperosmotic stress.
Discussion
This work is the first attempt to detect and understand the role of post-translational modifications of Kaiso. We found that Kaiso exists mainly as monoSUMOylated protein. At some physiological circumstances, cells are subjected to hyperosmotic stress that lead to their wrinkling and shrinkage. Cells respond to suddenly increased osmolarity by pumping in ions and water to compensate for external pressure. We have observed that Kaiso is quickly deSUMOylated in response to elevated concentrations of salt or sorbitol, which cause hyperosmotic shock. Thus, we think of Kaiso as a fast osmotic sensor. We see also that the deSUMOylation: (i) is reversible upon withdrawal of osmotic pressure and (ii) relaxes, returning Kaiso to almost fully SUMOylated form, if cells adapt to osmotic shock in 3–6 h and recover their shape. We can only speculate at this stage that Kaiso, a p120 catenin partner, somehow receives signals from the cytoskeleton, which becomes stressed upon osmotic shock, and participates in new transcriptional programs that are started in response to stress. One possible mechanism involves direct interactions between p120 and Kaiso that lead to the inhibition of DNA binding and Kaiso trafficking from nucleus to cytoplasm [39], where potential deSUMOylases act on the protein. Whether SUMOylation can regulate interactions of Kaiso with p120 and DNA should be addressed in further studies.
As we have shown here, Kaiso may be monoSUMOylated with SUMO-2,3. It is quite a rare case, and there is a striking example of monoSUMOylation with SUMO2, 3. This is the inhibitor of NFkB (IkBα). IkBα can SUMOylated and ubiquitinated at the same K21 lysine. After cells are stimulated, IkBα-SUMO1 cannot be ubiquitinated and remains stable, but IkBα-SUMO2 is polyubiquitinated and therefore exposed to proteasomal degradation [40, 41]. This example shows a possibility for monoSUMOylated proteins to be further modified. The same mechanism may work for Kaiso. However, up to now we do not have any evidence that Kaiso gene expression is regulated at the level of protein stability.
Previously published ChIP-seq analysis of Kaiso binding sites demonstrated a positive correlation between the binding of Kaiso and RNA expression level. This pioneering work hypothesized that Kaiso may attract transcriptional activators [42]. Now, based on our results, a positive correlation between Kaiso binding and promoter activity might be explained. A monoSUMOylated form of Kaiso is stable, but SUMOylation converts Kaiso from a repressor to an activator. We mapped the SUMOylation site within the BTB/POZ domain that is responsible for protein−protein interactions and dimerization [9, 15, 27]. BTB/POZ domain modifications may influence a spectrum of interacting proteins and consequently may have an impact on repression/activation properties of Kaiso. Tremendous progress in this direction would be achieved by raising monoclonal antibodies that specifically recognize monoSUMOylated Kaiso. Use of this reagent in ChIP-Seq and immunoprecipitation experiments would broaden our understanding of the role of SUMOylated Kaiso.
Transcriptional profiling of K42R mutant cells produced a cluster of deregulated genes that Gene Ontology assigned to the inflammatory response and blood pressure regulation (Supplementary Table 3). Our in vivo observation of increased hypertension in Kaiso-deficient animals that were kept on a high salt diet supported in vitro findings. Additionally, it was shown previously that hyperosmolarity stimulates the release of pro-inflammatory cytokines from immune and epithelial cells [43]. As seen from transcriptional profiling of K42R cells, the nonSUMOylated form of Kaiso reduced expression of IL26, CD40, TGFb1, and Il31RA (see full list of inflammatory genes in Supplementary Table 3). We suggest that deSUMOylated Kaiso is a compensatory mechanism that is activated in response to inflammation caused by changes in osmolarity. For instance, we show that Kaiso interacts with promoter of TRIM25, the gene that is active in HEK293 cells. When Kaiso is deSUMOylated either by hyperosmotic stress or gene editing (as in K42R cells), it downregulates TRIM25 transcription. Histones that are depleted with H3K4me3 and H3K27Ac and enriched with H3K9me3 forms a certain chromatin structure around TRIM25 promoter in K42R cells. It is this chromatin structure that makes TRIM25 stably epigenetically repressed in K42R cells even after knockdown of Kaiso by siRNA. We hypothesize that in some cellular processes, transient deSUMOylation of Kaiso after stress may lead to stable inactivation of particular genes by generating chromatin structures with histone marks that are unfavorable for transcription (Fig. 6E).
Our results also support previously published data on the role of TRIM25 in stress-induced apoptosis. Downregulation of TRIM25 resulted in massive apoptosis during early embryogenesis of medaka [38]. We demonstrated that stable downregulation of TRIM25 by Kaiso deSUMOylation in K42R cell lines is associated with increased apoptosis levels. Kaiso overexpression in KO cells downregulates TRIM25 transcription and increases the level of apoptosis. These data suggest that Kaiso may control osmolarity-induced apoptosis through regulation of TRIM25 activity.
Therefore, Kaiso can regulate TRIM25 expression by participating in the establishment of repressive chromatin marks. We suggest that SUMO signaling to Kaiso opens new modes of control upon transcription of Kaiso target genes and stress-induced apoptosis.
References
Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C. et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–79.
Geiss-Friedlander R, Melchior F. Concepts in SUMOylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56.
Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–8.
Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH. et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteom. 2008;7:132–44.
Wei W, Yang P, Pang J, Zhang S, Wang Y, Wang MH. et al. A stress-dependent SUMO4 SUMOylation of its substrate proteins. Biochem Biophys Res Commun. 2008;375:454–9.
Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML, Yang WM. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci Rep. 2016;6:26509.
Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms?. Genes Dev. 2004;18:2046–59.
Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–81.
Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34.
Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS. et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Mol Cell. 2012;46:335–50.
Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–9.
Zhigalova NA, Sokolov AS, Prokhortchouk EB, Zhenilo SV. [S100A3 is a new target gene of Kaiso in mouse skin]. Mol Biol. 2015;49:362–5.
Zhenilo SV, Musharova OS, Pokhorchuk EB. [Transcription factor Kaiso does not interact with hydroxymethylated DNA within CTGCNA sequence context]. Mol Biol. 2013;47:522–5.
Pierre CC, Longo J, Bassey-Archibong BI, Hallett RM, Milosavljevic S, Beatty L. et al. Methylation-dependent regulation of hypoxia inducible factor-1 alpha gene expression by the transcription factor Kaiso. Biochim Biophys Acta. 2015;1849:1432–41.
Koh DI, Han D, Ryu H, Choi WI, Jeon BN, Kim MK. et al. KAISO, a critical regulator of p53-mediated transcription of CDKN1A and apoptotic genes. Proc Natl Acad Sci USA. 2014;111:15078–83.
Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–8.
Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A. et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24
Martin Caballero I, Hansen J, Leaford D, Pollard S, Hendrich BD. The methyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouse embryogenesis, but play a redundant function in neural differentiation. PLoS ONE. 2009;4:e4315
Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G. et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–8.
Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D. et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208.
Feng M, Deerhake ME, Keating R, Thaisz J, Xu L, Tsaih SW. et al. Genetic analysis of blood pressure in 8 mouse intercross populations. Hypertension. 2009;54:802–9.
Shumskaya VS, Zhigalova NA, Prokhorchouk AV, Prokhorchouk EB. Distribution of Kaiso protein in mouse tissues. Histochem Cell Biol. 2014;143:29–43.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Hilgarth RS, Sarge KD. Detection of SUMOylated proteins. Methods Mol Biol. 2005;301:329–38.
Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23.
Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–9.
Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M. et al. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–50.
Ruzov A, Dunican DS, Prokhortchouk A, Pennings S, Stancheva I, Prokhortchouk E. et al. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development. 2004;131:6185–94.
Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ. et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–38.
Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23.
Capasso JM, Rivard CJ, Enomoto LM, Berl T. Chloride, not sodium, stimulates expression of the gamma subunit of Na/K-ATPase and activates JNK in response to hypertonicity in mouse IMCD3 cells. Proc Natl Acad Sci USA. 2003;100:6428–33.
Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T. et al. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc Natl Acad Sci USA. 1993;90:11117–21.
Ikeda K, Orimo A, Higashi Y, Muramatsu M, Inoue S. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000;472:9–13.
Thomson SD, Ali S, Pickles L, Taylor J, Pace PE, Lymboura M. et al. Analysis of estrogen-responsive finger protein expression in benign and malignant human breast. Int J Cancer. 2001;91:152–8.
Qin Y, Cui H, Zhang H. Overexpression of TRIM25 in lung cancer regulates tumor cell progression. Technol Cancer Res Treat. 2016;15:707–15.
Zhang P, Elabd S, Hammer S, Solozobova V, Yan H, Bartel F, et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene. 2015;34(46):5729–38.
Zhigalova NA, Zhenilo SV, Aitkhozhina DS, Prokhorchuk EB. [Bifunctional role of domain zinc fingers of methyl-DNA-binding protein Kaiso]. Mol Biol. 2010;44(2):263–74.
Aillet F, Lopitz-Otsoa F, Egana I, Hjerpe R, Fraser P, Hay RT, et al. Heterologous SUMO-2/3-ubiquitin chains optimize IkappaBalpha degradation and NF-kappaB activity. PLoS ONE. 2012;7(12):e51672.
Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2(2):233–9.
Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenet Chromat. 2013;6(1):13.
Brocker C, Thompson DC, Vasiliou V. The role of hyperosmotic stress in inflammation and disease. Biomol Concepts. 2012;3(4):345–64.
Acknowledgements
We thank Dr. B. Hendrich for providing MBD2−/−, Kaiso−/y fibroblasts cell line, Dr. R. Niedenthal for SUMO1-GFP, pcDNA3-UBC9 plasmids, Dr. P. Sergiev for HA-agarose. We thank Dr. A. Reynolds for providing with anti-Kaiso polyclonal rabbit. We thank Dr. I. Petrushanko and Dr. S. Chumakov for assistance in flow cytometry. We thank Dr. S. Salozhin for fruitful discussion of the work.
Funding
The work was supported by RNF 14-14-01202 and part of the work related with mice was funded by budget from project No. 0324-2018-0016 of the Federal Research Center “Institute of Cytology and Genetics” SB RAS.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by V. D'Angiolella.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhenilo, S., Deyev, I., Litvinova, E. et al. DeSUMOylation switches Kaiso from activator to repressor upon hyperosmotic stress. Cell Death Differ 25, 1938–1951 (2018). https://doi.org/10.1038/s41418-018-0078-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0078-7