Abstract
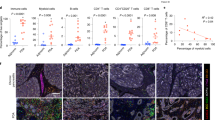
The tumor immune microenvironment (TIME) in ATC is a complex and diverse ecosystem. It is essential to have a comprehensive understanding to improve cancer treatment and prognosis. However, TIME of ATC and the dynamic changes with PTC has not been revealed at the single-cell level. Here, we performed an integrative single-cell analysis of PTC and ATC primary tumor samples. We found that immunosuppressive cells and molecules dominated the TIME in ATC. Specifically, the level of infiltration of exhausted CD8+ T cells, and M2 macrophages was increased, and that of NK cells, B cells, and M1 macrophages was decreased. The cytotoxicity of CD8+ T cells, γδT cells, and NK cells was decreased, and immune checkpoint molecules, such as LAG3, PD1, HAVCR2, and TIGIT were highly expressed in ATC. Our findings contribute to the comprehension of TIME in both PTC and ATC, offering insights into the immunosuppressive factors specifically associated with ATC. Targeting these immunosuppressive factors may activate the anti-tumor immune response in ATC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others

Data availability
All the single-cell and bulk RNA-sequencing data in this study are obtained from public datasets, which are available in the Gene Expression Omnibus (GEO) database.
Code availability
The codes generated during this study are available at the Github repository (https://github.com/fengchaobio/scRNA-seq_TIME_ATC).
References
Miranda-Filho A, Lortet-Tieulent J, Bray F, Cao B, Franceschi S, Vaccarella S, et al. Thyroid cancer incidence trends by histology in 25 countries: a population-based study. Lancet Diabetes Endocrinol. 2021;9:225–34.
Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, et al. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133.
Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388:2783–95.
Molinaro E, Romei C, Biagini A, Sabini E, Agate L, Mazzeo S, et al. Anaplastic thyroid carcinoma: from clinicopathology to genetics and advanced therapies. Nat Rev Endocrinol. 2017;13:644–60.
Jannin A, Escande A, Al Ghuzlan A, Blanchard P, Hartl D, Chevalier B, et al. Anaplastic thyroid carcinoma: an update. Cancers. 2022;14:1061.
Menicali E, Guzzetti M, Morelli S, Moretti S, Puxeddu E. Immune landscape of thyroid cancers: new insights. Front Endocrinol. 2021;11:1169.
Ahn J, Jin M, Song E, Ryu Y, Song DE, Kim S-Y, et al. Immune profiling of advanced thyroid cancers using fluorescent multiplex immunohistochemistry. Thyroid. 2021;31:61–7.
Giannini R, Moretti S, Ugolini C, Macerola E, Menicali E, Nucci N, et al. Immune profiling of thyroid carcinomas suggests the existence of two major phenotypes: an ATC-like and a PDTC-like. J Clin Endocrinol Metab. 2019;104:3557–75.
Cunha LL, Domingues GAB, Morari EC, Soares FA, Vassallo J, Ward LS. The immune landscape of the microenvironment of thyroid cancer is closely related to differentiation status. Cancer Cell Int. 2021;21:387.
Wang X, Peng W, Li C, Qin R, Zhong Z, Sun C. Identification of an immune-related signature indicating the dedifferentiation of thyroid cells. Cancer Cell Int. 2021;21:231.
Yin M, Di G, Bian M. Dysfunction of natural killer cells mediated by PD-1 and Tim-3 pathway in anaplastic thyroid cancer. Int Immunopharmacol. 2018;64:333–9.
Ryder M, Ghossein RA, Ricarte-Filho JCM, Knauf JA, Fagin JA. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr Relat Cancer. 2008;15:1069–74.
Luo Y, Yang Y-C, Ma B, Xu W-B, Liao T, Wang Y. Integrated analysis of novel macrophage related signature in anaplastic thyroid cancer. Endocrine. 2022;78:517–30.
Ugolini C, Basolo F, Proietti A, Vitti P, Elisei R, Miccoli P, et al. Lymphocyte and immature dendritic cell infiltrates in differentiated, poorly differentiated, and undifferentiated thyroid carcinoma. Thyroid. 2007;17:389–93.
Jung KY, Cho SW, Kim YA, Kim D, Oh B-C, Park DJ, et al. Cancers with higher density of tumor-associated macrophages were associated with poor survival rates. J Pathol Transl Med. 2015;49:318–24.
Qing W, Fang W-Y, Ye L, Shen L-Y, Zhang X-F, Fei X-C, et al. Density of tumor-associated macrophages correlates with lymph node metastasis in papillary thyroid carcinoma. Thyroid. 2012;22:905–10.
Schürch CM, Roelli MA, Forster S, Wasmer M-H, Brühl F, Maire RS, et al. Targeting CD47 in anaplastic thyroid carcinoma enhances tumor phagocytosis by macrophages and is a promising therapeutic strategy. Thyroid. 2019;29:979–92.
Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35–45.
Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775.e18–89.e18.
Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998.e20–1013.e20.
Lavin Y, Kobayashi S, Leader A, Amir E-AD, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750.e17–65.e17.
Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978.
Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342.e16–56.e16.
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829.e20–45.e20.
Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–72.
Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24:986.
Pu W, Shi X, Yu P, Zhang M, Liu Z, Tan L, et al. Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat Commun. 2021;12:6058.
Luo H, Xia X, Kim GD, Liu Y, Xue Z, Zhang L, et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv. 2021;7:eabf3657.
Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39:599–608.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573.e29–87.e29.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329.e4–37.e4.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
Chu Y. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat Med. 2023;29:1550–62.
Zheng Y, Chen Z, Han Y, Han L, Zou X, Zhou B, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. 2020;11:6268.
Ying L, Yan F, Meng Q, Yuan X, Yu L, Williams BRG, et al. Understanding immune phenotypes in human gastric disease tissues by multiplexed immunohistochemistry. J Transl Med. 2017;15:206.
Li J-P, Wu C-Y, Chen M-Y, Liu S-X, Yan S-M, Kang Y-F, et al. PD-1+CXCR5-CD4+ Th-CXCL13 cell subset drives B cells into tertiary lymphoid structures of nasopharyngeal carcinoma. J Immunother Cancer. 2021;9:e002101.
Yang Z, Wei X, Pan Y, Xu J, Si Y, Min Z, et al. A new risk factor indicator for papillary thyroid cancer based on immune infiltration. Cell Death Dis. 2021;12:1–14.
Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–9.
Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792.
Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–49.
Simoni Y, Becht E, Fehlings M, Loh CY, Koo S-L, Teng KWW, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–9.
Yuen GJ, Demissie E, Pillai S. B lymphocytes and cancer: a love–hate relationship. Trends Cancer. 2016;2:747–57.
Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–55.
Petitprez F, de Reyniès A, Keung EZ, Chen TW-W, Sun C-M, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–60.
Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–5.
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59.
Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92.
Park A, Yang Y, Lee Y, Kim MS, Park Y-J, Jung H, et al. Indoleamine-2,3-dioxygenase in thyroid cancer cells suppresses natural killer cell function by inhibiting NKG2D and NKp46 expression via STAT signaling pathways. J Clin Med. 2019;8:E842.
Silk JD, Hermans IF, Gileadi U, Chong TW, Shepherd D, Salio M, et al. Utilizing the adjuvant properties of CD1d-dependent NK T cells in T cell–mediated immunotherapy. J Clin Invest. 2004;114:1800–11.
Xie G, Dong H, Liang Y, Ham JD, Rizwan R, Chen J. CAR-NK cells: a promising cellular immunotherapy for cancer. eBioMedicine. 2020;59:102975.
Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017;14:662–74.
Yang Z, Yin L, Zeng Y, Li Y, Chen H, Yin S, et al. Diagnostic and prognostic value of tumor-infiltrating B cells in lymph node metastases of papillary thyroid carcinoma. Virchows Arch. 2021;479:947–59.
MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383.
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–47.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72.
Acknowledgements
We thank Dr. Guangchun Han and Tai Yanhong for the helpful discussions and Qinghai Provincial People’s Hospital for providing pathological tissue samples.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81572620) and Natural Science Foundation of Shandong Province (grant no. ZR2023MH370).
Author information
Authors and Affiliations
Contributions
Y.C. conceived this study. Y.C. and X.L. jointly supervised the study. C.F., Y.T., C.Y., L.W., and X.L. contributed to sample and information collection and data generation. Y.C. and C.F. supervised the bioinformatics data processing and analysis. C.F. performed bioinformatics data analysis. Y.T., C.Y., and L.W. assisted with data processing and analysis. X.L. and C.Y. performed IHC staining. Y.C., C.F., and X.L. wrote and revised the manuscript and all co-authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, C., Tao, Y., Yu, C. et al. Integrative single-cell transcriptome analysis reveals immune suppressive landscape in the anaplastic thyroid cancer. Cancer Gene Ther 30, 1598–1609 (2023). https://doi.org/10.1038/s41417-023-00663-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-023-00663-6