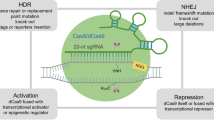
Abstract
Despite the development of new classes of targeted anti-cancer drugs, the curative treatment of metastatic solid tumors remains out of reach owing to the development of resistance to current chemotherapeutics. Although many mechanisms of drug resistance have been described, there is still a general lack of understanding of the many means by which cancer cells elude otherwise effective chemotherapy. The traditional strategy of isolating resistant clones in vitro, defining their mechanism of resistance, and testing to see whether these mechanisms play a role in clinical drug resistance is time-consuming and in many cases falls short of providing clinically relevant information. In this review, we summarize the use of CRISPR technology, including the promise and pitfalls, to generate libraries of cancer cells carrying sgRNAs that define novel mechanisms of resistance. The existing strategies using CRISPR knockout, activation, and inhibition screens, and combinations of these approaches are described. In addition, specialized approaches to identify more than one gene that may be contributing to resistance, as occurs in synthetic lethality, are described. Although these CRISPR-based approaches to cataloguing drug resistance genes in cancer cells are just beginning to be utilized, appropriately used they promise to accelerate understanding of drug resistance in cancer.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hakala MT, Zakrzewski SF, Nichol CA. Relation of folic acid reductase to amethopterin resistance in cultured mammalian cells. J Biol Chem. 1961;236:952–8.
Nunberg JH, Kaufman RJ, Schimke RT, Urlaub G, Chasin LA. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci USA. 1978;75:5553–6.
Dano K. Active outward transport of daunomycin in resistant Ehrlich ascites tumor cells. Biochim Biophys Acta. 1973;323:466–83.
Roninson IB, Chin JE, Choi KG, Gros P, Housman DE, Fojo A, et al. Isolation of human mdr DNA sequences amplified in multidrug-resistant KB carcinoma cells. Proc Natl Acad Sci USA. 1986;83:4538–42.
Ueda K, Cornwell MM, Gottesman MM, Pastan I, Roninson IB, Ling V, et al. The mdr1 gene, responsible for multidrug-resistance, codes for P-glycoprotein. Biochem Biophys Res Commun. 1986;141:956–62.
Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72.
Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7.
Sachse C, Echeverri CJ. Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene. 2004;23:8384–91.
Swanton C, Szallasi Z, Brenton JD, Downward J. Functional genomic analysis of drug sensitivity pathways to guide adjuvant strategies in breast cancer. Breast Cancer Res. 2008;10:214.
Yang H, Ren S, Yu S, Pan H, Li T, Ge S, et al. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int J Mol Sci. 2020;21:6460.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83.
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Spahn PN, Bath T, Weiss RJ, Kim J, Esko JD, Lewis NE, et al. PinAPL-Py: a comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci Rep. 2017;7:15854.
Kurata M, Rathe SK, Bailey NJ, Aumann NK, Jones JM, Veldhuijzen GW, et al. Using genome-wide CRISPR library screening with library resistant DCK to find new sources of Ara-C drug resistance in AML. Sci Rep. 2016;6:36199.
Barghout SH, Aman A, Nouri K, Blatman Z, Arevalo K, Thomas GE, et al. A genome-wide CRISPR/Cas9 screen in acute myeloid leukemia cells identifies regulators of TAK-243 sensitivity. JCI Insight. 2021;6:e141518.
Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Sci Transl Med. 2013;5:205rv1.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–76.
Wong AS, Choi GC, Cui CH, Pregernig G, Milani P, Adam M, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci USA. 2016;113:2544–9.
Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112:3570–5.
Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–74.
Qian S, Liang S, Yu H. Leveraging genetic interactions for adverse drug-drug interaction prediction. PLoS Comput Biol. 2019;15:e1007068.
le Sage C, Lawo S, Panicker P, Scales TME, Rahman SA, Little AS, et al. Dual direction CRISPR transcriptional regulation screening uncovers gene networks driving drug resistance. Sci Rep. 2017;7:17693.
Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, et al. Combined CRISPRi/a-based chemical genetic screens reveal that Rigosertib is a microtubule-destabilizing agent. Mol Cell. 2017;68:210–23.e6.
Palmer AC, Chidley C, Sorger PK. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. Elife. 2019;8:e50036.
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32.
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12.
Lin S, Larrue C, Scheidegger NK, Seong BKA, Dharia NV, Kuljanin M, et al. An in vivo CRISPR screening platform for prioritizing therapeutic targets in AML. Cancer Discov. 2022;12:432–49.
Merino D, Whittle JR, Vaillant F, Serrano A, Gong JN, Giner G, et al. Synergistic action of the MCL-1 inhibitor S63845 with current therapies in preclinical models of triple-negative and HER2-amplified breast cancer. Sci Transl Med. 2017;9:eaam7049.
Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–64.e20.
Wei L, Lee D, Law CT, Zhang MS, Shen J, Chin DW, et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10:4681.
Chen J, Huang Y, Tang Z, Li M, Ling X, Liao J, et al. Genome-scale CRISPR-Cas9 transcriptional activation screening in metformin resistance related gene of prostate cancer. Front Cell Dev Biol. 2020;8:616332.
Lau MT, Ghazanfar S, Parkin A, Chou A, Rouaen JR, Littleboy JB, et al. Systematic functional identification of cancer multi-drug resistance genes. Genome Biol. 2020;21:27.
Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao K, et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther. 2022;7:54.
Chen Y, Li L, Lan J, Cui Y, Rao X, Zhao J, et al. CRISPR screens uncover protective effect of PSTK as a regulator of chemotherapy-induced ferroptosis in hepatocellular carcinoma. Mol Cancer. 2022;21:11.
MacLeod G, Bozek DA, Rajakulendran N, Monteiro V, Ahmadi M, Steinhart Z, et al. Genome-wide CRISPR-Cas9 screens expose genetic vulnerabilities and mechanisms of temozolomide sensitivity in glioblastoma stem cells. Cell Rep. 2019;27:971–86.e9.
Szlachta K, Kuscu C, Tufan T, Adair SJ, Shang S, Michaels AD, et al. CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat Commun. 2018;9:4275.
Addgene. CRISPR Libraries. 2022. https://www.addgene.org/pooled-library/#crispr.
Agilent. CRISPR Libraries. 2022. https://www.agilent.com/en/product/crispr-cas/crispr-libraries.
GenScript. CRISPR gRNA Libraries. 2022. https://www.genscript.com/CRISPR-gRNA-library.html.
Colic M, Hart T. Common computational tools for analyzing CRISPR screens. Emerg Top Life Sci. 2021;5:779–88.
String Consortium. Protein-protein interaction networks, functional enrichment analysis. 2022. https://string-db.org/.
Enrichr. Analyze, Input data. 2022. https://maayanlab.cloud/Enrichr/.
GSEA. Molecular Signatures Database. 2022. https://www.gsea-msigdb.org/gsea/msigdb/.
Kaplan-Meier Plotter. What is the KM plotter? 2022. https://kmplot.com/analysis/.
Acknowledgements
We thank G. Leiman in the Laboratory of Cell Biology for editorial assistance.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute.
Author information
Authors and Affiliations
Contributions
Conception and design of the study: GA, HMW and AAB. Drafting the article: GA, HMW, AAB, CCL, MDS, RML, BAM, RWR and MMG. Revision and editing: GA, PM, RWR and MMG. All authors reviewed the final version before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alyateem, G., Wade, H.M., Bickert, A.A. et al. Use of CRISPR-based screens to identify mechanisms of chemotherapy resistance. Cancer Gene Ther 30, 1043–1050 (2023). https://doi.org/10.1038/s41417-023-00608-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-023-00608-z
This article is cited by
-
From pre-clinical to translational brain metastasis research: current challenges and emerging opportunities
Clinical & Experimental Metastasis (2024)