Abstract
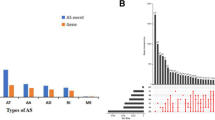
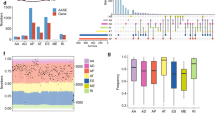
Alternative splicing (AS) is a gene regulatory mechanism that drives protein diversity and dysregulation of AS plays a significant role in tumorigenesis. This study aimed to develop a prognostic signature based on AS and elucidate the role in tumor immune microenvironment (TIME) in clear cell renal cell carcinoma (ccRCC). The prognosis-related AS events were analyzed by univariate Cox regression analysis. Gene set enrichment analyses (GSEA) were performed for functional annotation. Prognostic signatures were identified and validated using univariate and multivariate Cox regression, LASSO regression, Kaplan–Meier survival analysis, and proportional hazards model. The context of TIME in ccRCC was also analyzed. Gene and protein expression data of C4orf19 were obtained from ONCOMINE website and Human Protein Altas. Splicing factors (SFs) regulatory networks were visualized. 4431 survival-related AS events in ccRCC were screened. Based on splicing subtypes, eight AS prognostic signatures were constructed. A nomogram with good prognostic prediction was generated. Furthermore, the prognostic signatures were significantly correlated with TIME diversity and immune checkpoint inhibitor (ICI)-related genes. C4orf19 was the only gene whose expression levels were downregulated among the prognostic AS-related genes, which is considered as a promising prognostic factor in ccRCC. Potential functions of SFs were determined by splicing regulatory networks. In our study, AS patterns of novel indicators for prognostic prediction of ccRCC were explored. The AS-SF networks provide information of regulatory mechanisms. Players of AS events related to TIME were investigated, which contribute to prognosis monitoring of ccRCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study.
References
Motzer RJ, Chang SS, Jonasch E, Choueiri TK, Hancock SL, Lin DW, et al. Kidney cancer, version 3.2015. Pract Guide. 2015;13:151–9.
Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34:193–205.
Zuo Y, Zhang L, Tang W, Tang W. Identification of prognosis-related alternative splicing events in kidney renal clear cell carcinoma. J Cell Mol Med. 2019;23:7762–72.
Weber J. Immune checkpoint proteins: a new therapeutic paradigm for cancer–preclinical background: CTLA-4 and PD-1 blockade. Semin Oncol. 2010;37:430–9.
Kruger S, Ilmer M, Kobold S, Cadilha BL, Endres S, Ormanns S, et al. Advances in cancer immunotherapy 2019 - latest trends. J Exp Clin Cancer Res. 2019;38:268.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–28.
Montes M, Sanford BL, Comiskey DF, Chandler DS. RNA splicing and disease: animal models to therapies. Trends Genet. 2019;35:68–87.
Frankiw L, Baltimore D, Li G. Alternative mRNA splicing in cancer immunotherapy. Nat Rev Immunol. 2019;19:675–87.
Wu HY, Peng ZG, He RQ, Luo B, Ma J, Hu XH, et al. Prognostic index of aberrant mRNA splicing profiling acts as a predictive indicator for hepatocellular carcinoma based on TCGA SpliceSeq data. Int J Oncol. 2019;55:425–38.
Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23:282–96. e4
Kouyama Y, Masuda T, Fujii A, Ogawa Y, Sato K, Tobo T, et al. Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 2019;110:3132–44.
Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–86.
Zhou M, Diao Z, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383–94.
Wang C, Zheng M, Wang S, Nie X, Guo Q, Gao L, et al. Whole genome analysis and prognostic model construction based on alternative splicing events in endometrial cancer. Biomed Res Int. 2019;2019:2686875.
Meng T, Huang R, Zeng Z, Huang Z, Yin H, Jiao C, et al. Identification of prognostic and metastatic alternative splicing signatures in kidney renal clear cell carcinoma. Front Bioeng Biotechnol. 2019;7:270.
Xiao L, Zou G, Cheng R, Wang P, Ma K, Cao H, et al. Alternative splicing associated with cancer stemness in kidney renal clear cell carcinoma. BMC Cancer. 2021;21:703.
Ryan MC, Cleland J, Kim R, Wong WC, Weinstein JN. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–7.
Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381–97.
Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol. 2017;14:203–20.
Xu Q, Xu H, Deng R, Li N, Mu R, Qi Z, et al. Immunological significance of prognostic alternative splicing signature in hepatocellular carcinoma. Cancer Cell Int. 2021;21:190.
Kim JE, Patel MA, Mangraviti A, Kim ES, Theodros D, Velarde E, et al. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res. 2017;23:124–36.
Zhai L, Ladomersky E, Lenzen A, Nguyen B, Patel R, Lauing KL, et al. IDO1 in cancer: a Gemini of immune checkpoints. Cell Mol Immunol. 2018;15:447–57.
Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655–68.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Heidegger I, Pircher A, Pichler R. Targeting the tumor microenvironment in renal cell cancer biology and therapy. Front Oncol. 2019;9:490.
Finke JH, Rayman P, Edinger M, Tubbs RR, Stanley J, Klein E, et al. Characterization of a human renal cell carcinoma specific cytotoxic CD8+ T cell line. J Immunother. 1992;11:1–11.
Fyfe BG, Fisher RI, Rosenberg SA, Sznol M, Parkinson DR, Louie AC. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. Clin Trial. 1995;13:688–96.
Figlin RA, Belldegrun A, Moldawer N, Zeffren J, deKernion J. Concomitant administration of recombinant human interleukin-2 and recombinant interferon alfa-2A: an active outpatient regimen in metastatic renal cell carcinoma. Clin Trial. 1992;10:414–21.
Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V, et al. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Pr Guide. 2019;30:706–20.
Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V, et al. The role of immunotherapy in solid tumors: report from the Campania Society of Oncology Immunotherapy (SCITO) meeting, Naples 2014. J Transl Med. 2014;12:291.
Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:681–700.
Hu J, Chen Z, Bao L, Zhou L, Hou Y, Liu L, et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes. Mol Ther. 2020;28:1658–72.
Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63.
Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–26.
Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–49. e18
Lang ZQ, Wu YQ, Pan XB, Qu GM, Zhang TG. The identification of multifocal breast cancer-associated long non-coding RNAs. Eur Rev Med Pharm Sci. 2017;21:5648–54.
Author information
Authors and Affiliations
Contributions
ZW, LP, KL, and PZ designed and supervised the study. ZW, LZ, LL, YS, and LP analyzed the data and wrote the original draft. LP, GG, and JS edited the draft. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
JS’ conflicts can be found at https://www.nature.com/onc/editors. GG is Editor in Chief in Cancer Gene Therapy and the Founder and Chief Scientific Advisor of Stingray Bio. None are relevant here. Other authors do not declare conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Z., Zhu, L., Li, K. et al. Alternative splicing events in tumor immune infiltration in renal clear cell carcinomas. Cancer Gene Ther 29, 1418–1428 (2022). https://doi.org/10.1038/s41417-022-00426-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-022-00426-9