Abstract
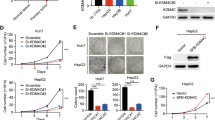
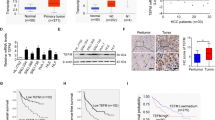
Hepatocellular carcinoma (HCC) is one of the deadliest cancer types worldwide. The centromere proteins (CENPs) are critical for the mitosis-related protein complex and are involved in kinetochore assembly and spindle checkpoint signaling during mitosis. However, the clinical significance of CENPs in the recurrence and progression of HCC remains poorly understood. Here, we examined the expression of all CENPs and their association with recurrence and survival of HCC patients using the global gene expression profile dataset established in our laboratory. The effect of silencing CENPF on cell viability, migration, and epithelial-mesenchymal transition (EMT) were detected using CCK-8, transwell, and western blot, respectively. RT-qPCR and western blot were performed to confirm the silencing of CENPF and the relationship between STAT5A and CENPF, while tumorigenesis was tested using the HCC Huh7 xenograft mouse model. Most of the CENPs is overexpressed in HCC, and overexpression of CENPF was significantly associated with the poor survival of HCC patients. CENPF promoted HCC cell lines migration and EMT progression. Knockdown CENPF inhibited cell growth activity against human HCC cells in vitro and xenograft tumors in vivo. Bioinformatics analysis revealed that CENPF genes are enriched in the cell cycle. Silencing CENPF arrested cell cycle at the G2/M phase and inhibited Cyclin B1 and Cyclin E1 expressions. Meanwhile, silencing CENPF prohibited phosphorylation of ERK and the expression of NEK2. Additionally, we found that STAT5A down-regulated CENPF expression and inhibited cancer cell growth viability. In conclusion, our data suggested that CENPF could be potentially developed into a theranostic biomarker to tackle HCC progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Chen H-J, Hu M-H, Xu F-G, Xu H-J, She J-J, Xia H-P. Understanding the inflammation-cancer transformation in the development of primary liver cancer. Hepatoma Res. 2018;4:29.
Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu X, et al. Liver cancer incidence and mortality in China: Temporal trends and projections to 2030. Chin J cancer Res = Chung-kuo yen cheng yen chiu. 2018;30:571–9.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14.
Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115.
Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol cell Biol. 2011;12:320–32.
Egloff AM, Vella LA, Finn OJ. Cyclin B1 and Other Cyclins as Tumor Antigens in Immunosurveillance and Immunotherapy of Cancer. Cancer Res. 2006;66:6–9.
Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK et al. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019;8:1118.
Di Agostino S, Rossi P, Geremia R, Sette C. The MAPK pathway triggers activation of Nek2 during chromosome condensation in mouse spermatocytes. Development. 2002;129:1715–27.
Wen S, Liu Y, Yang M, Yang K, Huang J, Feng D. Increased NEK2 in hepatocellular carcinoma promotes cancer progression and drug resistance by promoting PP1/Akt and Wnt activation. Oncol Rep. 2016;36:2193–9.
Shahid M, Kim M, Lee MY, Yeon A, You S, Kim HL, et al. Downregulation of CENPF Remodels Prostate Cancer Cells and Alters Cellular Metabolism. Proteomics. 2019;19:e1900038.
Dai Y, Liu L, Zeng T, Zhu YH, Li J, Chen L, et al. Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma. Biochemical biophysical Res Commun. 2013;436:711–8.
Cheng Y, Hong M, Cheng B. Identified differently expressed genes in renal cell carcinoma by using multiple microarray datasets running head: differently expressed genes in renal cell carcinoma. Eur Rev Med Pharm Sci. 2014;18:1033–40.
Michalak M, Warnken U, Schnolzer M, Gabius HJ, Kopitz J. Detection of malignancy-associated phosphoproteome changes in human colorectal cancer induced by cell surface binding of growth-inhibitory galectin-4. IUBMB life. 2019;71:364–75.
Li R, Wang X, Zhao X, Zhang X, Chen H, Ma Y, et al. Centromere protein F and Forkhead box M1 correlation with prognosis of non-small cell lung cancer. Oncol Lett. 2020;19:1368–74.
Sun J, Huang J, Lan J, Zhou K, Gao Y, Song Z, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer cell Int. 2019;19:264.
Shi J, Zhang P, Liu L, Min X, Xiao Y. Weighted gene coexpression network analysis identifies a new biomarker of CENPF for prediction disease prognosis and progression in nonmuscle invasive bladder cancer. Mol Genet Genom Med. 2019;7:e982.
Zhou CJ, Wang XY, Han Z, Wang DH, Ma YZ, Liang CG. Loss of CENPF leads to developmental failure in mouse embryos. Cell cycle. 2019;18:2784–99.
Liu ZK, Zhang RY, Yong YL, Zhang ZY, Li C, Chen ZN, et al. Identification of crucial genes based on expression profiles of hepatocellular carcinomas by bioinformatics analysis. PeerJ. 2019;7:e7436.
Feitelson MA, Arzumanyan A, Kulathinal RJ, Blain SW, Holcombe RF, Mahajna J, et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin Cancer Biol. 2015;35:S25–s54. Suppl(Suppl)
Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, et al. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011;6:e17794.
Li Z, Liu B, Jin W, Wu X, Zhou M, Liu VZ et al. hDNA2 nuclease/helicase promotes centromeric DNA replication and genome stability. The EMBO journal 2018;37:e96729.
Holen EIH KD, Schelman WR, Kirby LC, Johnson BM, Botbyl JD, Grilley-Olson JE, et al. Phase I first-in-human study of the centromere-associated protein E (CENP-E) inhibitor GSK923295 in patients with advanced solid tumors (study CPE107602). J Clin Oncol. 2010;28:3012–3012. May 2015_suppl
Bennett A, Bechi B, Tighe A, Thompson S, Procter DJ, Taylor SS. Cenp-E inhibitor GSK923295: Novel synthetic route and use as a tool to generate aneuploidy. Oncotarget. 2015;6:20921–32.
Cao JY, Liu L, Chen SP, Zhang X, Mi YJ, Liu ZG, et al. Prognostic significance and therapeutic implications of centromere protein F expression in human nasopharyngeal carcinoma. Mol cancer. 2010;9:237.
Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin cancer Res: CR. 2019;38:188.
Lin SC, Kao CY, Lee HJ, Creighton CJ, Ittmann MM, Tsai SJ, et al. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat Commun. 2016;7:11418.
Yu JH, Zhu BM, Riedlinger G, Kang K, Hennighausen L. The liver-specific tumor suppressor STAT5 controls expression of the reactive oxygen species-generating enzyme NOX4 and the proapoptotic proteins PUMA and BIM in mice. Hepatology. 2012;56:2375–86.
Yu JH, Zhu BM, Wickre M, Riedlinger G, Chen W, Hosui A, et al. The transcription factors signal transducer and activator of transcription 5A (STAT5A) and STAT5B negatively regulate cell proliferation through the activation of cyclin-dependent kinase inhibitor 2b (Cdkn2b) and Cdkn1a expression. Hepatology. 2010;52:1808–18.
Jiang Y, Tao Y, Zhang X, Wei X, Li M, He X, et al. Loss of STAT5A promotes glucose metabolism and tumor growth through miRNA-23a-AKT signaling in hepatocellular carcinoma. Mol Oncol. 2020;15:710–24.
Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging. 2011;3:192–222.
Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma (Review). Oncol Lett. 2017;13:1041–7.
Liang Y, Chen J, Yu Q, Ji T, Zhang B, Xu J, et al. Phosphorylated ERK is a potential prognostic biomarker for Sorafenib response in hepatocellular carcinoma. Cancer Med. 2017;6:2787–95.
Acknowledgements
This work was supported by grants from the National Medical Research Council (NMRC) of Singapore, The Recruitment Program of Overseas High-Level Young Talents, National Natural Science Foundation of China (82072739), “Innovative and Entrepreneurial Team” (No. (2018)2015), Science and Technology Grant (BE2019758) and the Six Talent Peaks Project (TD-SWYY-007) of Jiangsu Province and High-Level Talents Program of Nanjing Medical University, and Chinese Foundation for Hepatitis Prevention and Control-Tian Qing Liver Disease Research Fund (TQGB20190164, TQGB20200139). The China Scholarship Council [201908320572 to H.C.] and Nanjing Medical University Scholarship [C124 to H.C.].
Author information
Authors and Affiliations
Contributions
H.C., K.H., and H.X. conceived the idea and designed the experiments. H.C. and H.X. carried out the experiments. F.W., F.X., and G.Z. contributed to the clinical samples and analysis. G.W., M.D., and A.D. help the experimental and analysis. H.C. wrote the paper with input from all authors. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, H., Wu, F., Xu, H. et al. Centromere protein F promotes progression of hepatocellular carcinoma through ERK and cell cycle-associated pathways. Cancer Gene Ther 29, 1033–1042 (2022). https://doi.org/10.1038/s41417-021-00404-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00404-7
This article is cited by
-
Aberrant activation of five embryonic stem cell-specific genes robustly predicts a high risk of relapse in breast cancers
BMC Genomics (2023)
-
ATAD2 is a driver and a therapeutic target in ovarian cancer that functions by upregulating CENPE
Cell Death & Disease (2023)
-
Integrated pan-cancer analysis of centromere protein F and experimental verification of its role and clinical significance in cholangiocarcinoma
Functional & Integrative Genomics (2023)