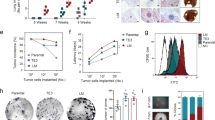
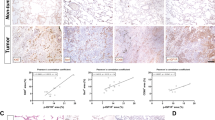
Abstract
Triple-negative breast cancer (TNBCs) display lung metastasis tropism. However, the mechanisms underlying this organ-specific pattern remains to be elucidated. We sought to evaluate the utility of blocking extravasation to prevent lung metastasis. To identify potential geminin overexpression-controlled genetic drivers that promote TNBC tumor homing to lungs, we used the differential/suppression subtractive chain (D/SSC) technique. A geminin overexpression-induced lung metastasis gene signature consists of 24 genes was discovered. We validated overexpression of five of these genes (LGR5, HAS2, CDH11, NCAM2, and DSC2) in worsening lung metastasis-free survival in TNBC patients. Our data demonstrate that LGR5-induced β-catenin signaling and stemness in TNBC cells are geminin-overexpression dependent. They also demonstrate for the first-time expression of RSPO2 in mouse lung tissue only and exacerbation of its secretion in the circulation of mice that develop geminin overexpressing/LGR5+-TNBC lung metastasis. We identified a novel extravasation receptor complex, consists of CDH11, CD44v6, c-Met, and AXL on geminin overexpressing/LGR5+-TNBC lung metastatic precursors, inhibition of any of its receptors prevented geminin overexpressing/LGR5+-TNBC lung metastasis. Overall, we propose that geminin overexpression in normal mammary epithelial (HME) cells promotes the generation of TNBC metastatic precursors that home specifically to lungs by upregulating LGR5 expression and promoting stemness, intravasation, and extravasation in these precursors. Circulating levels of RSPO2 and OPN can be diagnostic biomarkers to improve risk stratification of metastatic TNBC to lungs, as well as identifying patients who may benefit from therapy targeting geminin alone or in combination with any member of the newly discovered extravasation receptor complex to minimize TNBC lung metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author [WeS].
References
Gennari A, Conte P, Rosso R, Orlandini C, Bruzzi P. Survival of metastatic breast carcinoma patients over a 20-year period: a retrospective analysis based on individual patient data from six consecutive studies. Cancer. 2005;104:1742–50.
Jin L, Han B, Siegel E, Cui Y, Giuliano A, Cui X. Breast cancer lung metastasis: molecular biology and therapeutic implications. Cancer Biol Ther. 2018;19:858–68.
Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92.
Castagnoli L, Cancila V, Cordoba-Romero SL, Faraci S, Talarico G, Belmonte B, et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38:4047–60.
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7.
Nakata S, Campos B, Bageritz J, Bermejo JL, Becker N, Engel F, et al. LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol. 2013;23:60–72.
Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–7.
Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, Whitsett JA. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Dev. 2008;135:1049–58.
Xu J, Chen Y, Huo D, Khramtsov A, Khramtsova G, Zhang C, et al. β-catenin regulates c-Myc and CDKN1A expression in breast cancer cells. Mol Carcinog. 2016;55:431–9.
de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–16.
Chartier C, Raval J, Axelrod F, Bond C, Cain J, Dee-Hoskins C, et al. Therapeutic targeting of tumor-derived R-spondin attenuates β-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res. 2016;76:713–23.
Moumen M, Chiche A, Decraene C, Petit V, Gandarillas A, Deugnier MA, et al. Myc is required for β-catenin-mediated mammary stem cell amplification and tumorigenesis. Mol Cancer. 2013;12:132.
Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–74.
Sato K, Uehara T, Iwaya M, Nakajima T, Miyagawa Y, Suga T, et al. Correlation of clinicopathological features and LGR5 expression in colon adenocarcinoma. Ann Diagn Pathol. 2019;40:161–5.
Martinez C, Churchman M, Freeman T, Ilyas M. Osteopontin provides early proliferative drive and may be dependent upon aberrant c-myc signalling in murine intestinal tumours. Exp Mol Pathol. 2010;88:272–7.
Wang X, Chao L, Ma G, Chen L, Tian B, Zang Y, et al. Increased expression of osteopontin in patients with triple-negative breast cancer. Eur J Clin Invest. 2008;38:438–46.
Inglis DJ, Lavranos TC, Beaumont DM, Leske AF, Brown CK, Hall AJ, et al. The vascular disrupting agent BNC105 potentiates the efficacy of VEGF and mTOR inhibitors in renal and breast cancer. Cancer Biol Ther. 2014;15:1552–60.
Rittling SR, Novick KE. Osteopontin expression in mammary gland development and tumorigenesis. Cell Growth Differ. 1997;8:1061–9.
Zhou Y, Xia L, Wang H, Oyang L, Su M, Liu Q, et al. Cancer stem cells in progression of colorectal cancer. Oncotarget. 2018;9:33403–15.
Heldin P, Basu K, Kozlova I, Porsch H. HAS2 and CD44 in breast tumorigenesis. Adv Cancer Res. 2014;123:211–29.
Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11:64.
Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883.
Hu J, Li G, Zhang P, Zhuang X, Hu G. A CD44v(+) subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017;8:e2679.
Bellerby R, Smith C, Kyme S, Gee J, Günthert U, Green A, et al. Overexpression of specific CD44 isoforms is associated with aggressive cell features in acquired endocrine resistance. Front Oncol. 2016;6:145.
Blanchard Z, Malik R, Mullins N, Maric C, Luk H, Horio D, et al. Geminin overexpression induces mammary tumors via suppressing cytokinesis. Oncotarget. 2011;2:1011–27.
Blanchard Z, Mullins N, Ellipeddi P, Lage JM, McKinney S, El-Etriby R, et al. Geminin overexpression promotes imatinib sensitive breast cancer: a novel treatment approach for aggressive breast cancers, including a subset of triple negative. PLoS ONE. 2014;9:e95663.
Ananthula S, Sinha A, El Gassim M, Batth S, Marshall GD Jr., Gardner LH, et al. Geminin overexpression-dependent recruitment and crosstalk with mesenchymal stem cells enhance aggressiveness in triple negative breast cancers. Oncotarget. 2016;7:20869–89.
Gardner L, Malik R, Shimizu Y, Mullins N, ElShamy WM. Geminin overexpression prevents the completion of topoisomerase IIalpha chromosome decatenation, leading to aneuploidy in human mammary epithelial cells. Breast Cancer Res. 2011;13:R53.
Ryan D, Koziol J, ElShamy WM. Targeting AXL and RAGE to prevent geminin overexpression-induced triple-negative breast cancer metastasis. Sci Rep. 2019;9:19150.
D’Alfonso TM, Hannah J, Chen Z, Liu Y, Zhou P, Shin SJ. Axl receptor tyrosine kinase expression in breast cancer. J Clin Pathol. 2014;67:690–6.
Lozneanu L, Pinciroli P, Ciobanu DA, Carcangiu ML, Canevari S, Tomassetti A, et al. Computational and immunohistochemical analyses highlight AXL as a potential prognostic marker for ovarian cancer patients. Anticancer Res. 2016;36:4155–63.
Nakuci E, Xu M, Pujana MA, Valls J, Elshamy WM. Geminin is bound to chromatin in G2/M phase to promote proper cytokinesis. Int J Biochem Cell Biol. 2006;38:1207–20.
Pedroza M, Welschhans RL, Agarwal SK. Targeting of cadherin-11 decreases skin fibrosis in the tight skin-1 mouse model. PLoS ONE. 2017;12:e0187109.
Nasser MW, Wani NA, Ahirwar DK, Powell CA, Ravi J, Elbaz M, et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Res. 2015;75:974–85.
Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Li L, Techel D, Gretz N, Hildebrandt A. A novel transcriptome subtraction method for the detection of differentially expressed genes in highly complex eukaryotes. Nucleic Acids Res. 2005;33:e136.
Røsok Ø, Sioud M. Discovery of differentially expressed genes: technical considerations. Methods Mol Biol. 2007;360:115–29.
Liu BH, Goh CH, Ooi LL, Hui KM. Identification of unique and common low abundance tumour-specific transcripts by suppression subtractive hybridization and oligonucleotide probe array analysis. Oncogene. 2008;27:4128–36.
Luo JH, Puc JA, Slosberg ED, Yao Y, Bruce JN, Wright TC Jr., et al. Differential subtraction chain, a method for identifying differences in genomic DNA and mRNA. Nucleic Acids Res. 1999;27:e24.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Leelatian N, Doxie DB, Greenplate AR, Sinnaeve J, Ihrie RA, Irish JM. Preparing viable single cells from human tissue and tumors for cytomic analysis. Curr Protoc Mol Biol. 2017;118:25c.1.1–25c.1.23.
Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 2010;5:e9370.
Wang DY, Jiang Z, Ben-David Y, Woodgett JR, Zacksenhaus E. Molecular stratification within triple-negative breast cancer subtypes. Sci Rep. 2019;9:19107.
Fu F, Xiao XI, Zhang T, Zou Q, Chen Z, Pei L, et al. Expression of receptor protein tyrosine phosphatase zeta is a risk factor for triple negative breast cancer relapse. Biomed Rep. 2016;4:167–72.
Gerashchenko TS, Zavyalova MV, Denisov EV, Krakhmal NV, Pautova DN, Litviakov NV, et al. Intratumoral morphological heterogeneity of breast cancer as an indicator of the metastatic potential and tumor chemosensitivity. Acta Nat. 2017;9:56–67.
Li P, Xiang T, Li H, Li Q, Yang B, Huang J, et al. Hyaluronan synthase 2 overexpression is correlated with the tumorigenesis and metastasis of human breast cancer. Int J Clin Exp Pathol. 2015;8:12101–14.
Sagara A, Igarashi K, Otsuka M, Karasawa T, Gotoh N, Narita M, et al. Intrinsic resistance to 5-fluorouracil in a brain metastatic variant of human breast cancer cell line, MDA-MB-231BR. PLoS ONE. 2016;11:e0164250.
Takahashi S, Kato K, Nakamura K, Nakano R, Kubota K, Hamada H. Neural cell adhesion molecule 2 as a target molecule for prostate and breast cancer gene therapy. Cancer Sci. 2011;102:808–14.
Wang W, Meng Y, Dong B, Dong J, Ittmann MM, Creighton CJ, et al. A versatile tumor gene deletion system reveals a crucial role for FGFR1 in breast cancer metastasis. Neoplasia. 2017;19:421–8.
Zhao J, Ni H, Ma Y, Dong L, Dai J, Zhao F, et al. TIP30/CC3 expression in breast carcinoma: relation to metastasis, clinicopathologic parameters, and P53 expression. Hum Pathol. 2007;38:293–8.
Han Y, Xue X, Jiang M, Guo X, Li P, Liu F, et al. LGR5, a relevant marker of cancer stem cells, indicates a poor prognosis in colorectal cancer patients: a meta-analysis. Clin Res Hepatol Gastroenterol. 2015;39:267–73.
Hou MF, Chen PM, Chu PY. LGR5 overexpression confers poor relapse-free survival in breast cancer patients. BMC Cancer. 2018;18:219.
Dey N, Barwick BG, Moreno CS, Ordanic-Kodani M, Chen Z, Oprea-Ilies G, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537.
Chu JE, Xia Y, Chin-Yee B, Goodale D, Croker AK, Allan AL. Lung-derived factors mediate breast cancer cell migration through CD44 receptor-ligand interactions in a novel ex vivo system for analysis of organ-specific soluble proteins. Neoplasia. 2014;16:180–91.
ElShamy WM, Sinha A, Said N. Aggressiveness niche: can it be the foster ground for cancer metastasis precursors? Stem Cells Int. 2016;2016:4829106.
Sinha A, Paul BT, Sullivan LM, Sims H, El Bastawisy A, Yousef HF, et al. BRCA1-IRIS overexpression promotes and maintains the tumor initiating phenotype: implications for triple negative breast cancer early lesions. Oncotarget. 2017;8:10114–35.
Chen Z, Xue C. G-protein-coupled receptor 5 (LGR5) overexpression activates β-catenin signaling in breast cancer cells via protein kinase A. Med Sci Monit Basic Res. 2019;25:15–25.
Yang L, Tang H, Kong Y, Xie X, Chen J, Song C, et al. LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/β-catenin signaling. Stem Cells. 2015;33:2913–24.
Wang J, Li M, Chen D, Nie J, Xi Y, Yang X, et al. Expression of C-myc and β-catenin and their correlation in triple negative breast cancer. Minerva Med. 2017;108:513–7.
Cao HZ, Liu XF, Yang WT, Chen Q, Zheng PS. LGR5 promotes cancer stem cell traits and chemoresistance in cervical cancer. Cell Death Dis. 2017;8:e3039.
Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Dev. 2014;141:2206–15.
Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–5.
Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–26.
Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, et al. Gas6/Axl axis contributes to chemoresistance and metastasis in breast cancer through Akt/GSK-3beta/beta-catenin signaling. Theranostics. 2016;6:1205–19.
Hutchinson KE, Yost SE, Chang CW, Johnson RM, Carr AR, McAdam PR, et al. Comprehensive profiling of poor-risk paired primary and recurrent triple-negative breast cancers reveals immune phenotype shifts. Clin Cancer Res. 2020;26:657–68.
Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–67.
Sheng L, Leshchyns’ka I, Sytnyk V. Neural cell adhesion molecule 2 promotes the formation of filopodia and neurite branching by inducing submembrane increases in Ca2+ levels. J Neurosci. 2015;35:1739–52.
Satriyo PB, Bamodu OA, Chen JH, Aryandono T, Haryana SM, Yeh CT et al. Cadherin 11 inhibition downregulates β-catenin, deactivates the canonical WNT signalling pathway and suppresses the cancer stem cell-like phenotype of triple negative breast cancer. J Clin Med. 2019;8:148.
Roycroft A, Mayor R. Molecular basis of contact inhibition of locomotion. Cell Mol Life Sci. 2016;73:1119–30.
Culhane AC, Quackenbush J. Confounding effects in “A six-gene signature predicting breast cancer lung metastasis”. Cancer Res. 2009;69:7480–5.
van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6.
Coussy F, Lallemand F, Vacher S, Schnitzler A, Chemlali W, Caly M, et al. Clinical value of R-spondins in triple-negative and metaplastic breast cancers. Br J Cancer. 2017;116:1595–603.
Neophytou CM, Kyriakou TC, Papageorgis P. Mechanisms of metastatic tumor dormancy and implications for cancer therapy. Int J Mol Sci. 2019;20:6158.
Fujisaki T, Tanaka Y, Fujii K, Mine S, Saito K, Yamada S, et al. CD44 stimulation induces integrin-mediated adhesion of colon cancer cell lines to endothelial cells by up-regulation of integrins and c-Met and activation of integrins. Cancer Res. 1999;59:4427–34.
Zhang Z, Dong Z, Lauxen IS, Filho MS, Nor JE. Endothelial cell-secreted EGF induces epithelial to mesenchymal transition and endows head and neck cancer cells with stem-like phenotype. Cancer Res. 2014;74:2869–81.
Qiao GL, Song LN, Deng ZF, Chen Y, Ma LJ. Prognostic value of CD44v6 expression in breast cancer: a meta-analysis. Onco Targets Ther. 2018;11:5451–7.
Acknowledgements
Wael is Dr. Lawrence & Mrs. Bo Hing Chan Tseu, American Cancer Society Research Scholar.
Funding
In part by grant # RSG-09-275-01 from the American Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sami, E., Bogan, D., Molinolo, A. et al. The molecular underpinning of geminin-overexpressing triple-negative breast cancer cells homing specifically to lungs. Cancer Gene Ther 29, 304–325 (2022). https://doi.org/10.1038/s41417-021-00311-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-021-00311-x
This article is cited by
-
TAM family kinases as therapeutic targets at the interface of cancer and immunity
Nature Reviews Clinical Oncology (2023)
-
Varying outcomes of triple-negative breast cancer in different age groups–prognostic value of clinical features and proliferation
Breast Cancer Research and Treatment (2022)
-
Gene expression profiling and protein–protein interaction analysis reveals the dynamic role of MCM7 in Alzheimer's disorder and breast cancer
3 Biotech (2022)