Abstract
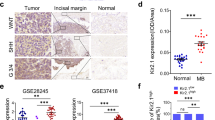
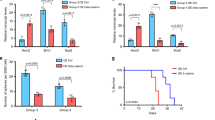
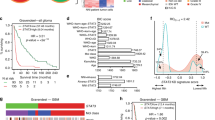
Medulloblastoma (MB) is the most frequent malignant brain tumor in children and it is subgrouped into 4 entities (SHH, WNT, Group 3, and Group 4). Molecular pathways involved in these different subgroups still are evolving and can be of clinical relevance to therapy. The YAP1-CTGF axis is known to regulate cell proliferation, differentiation, and cell death; however, its role in MB is poorly explored. We aimed to investigate the role of YAP1 gene in the MB SHH cell line DAOY and evaluate cell proliferation, doubling time and 3D spheroids invasion and its consequence on CTGF regulation. We assessed CTGF expression from 22 children with MB. Lastly, we validated our findings through in silico analysis in large cohorts dataset of patients. We observed an increased invasion rate of DAOY cells and CTGF downregulation under YAP1 knockdown (p < 0.0001). Additionally CTGF is overexpressed in MB with extensive nodularity subtype and an indicative of higher survival rates in pediatric MB (p < 0.05). Interestingly, no difference of CTGF expression was observed between molecular subgroups. These results provide new evidence ofCTGF as a potential prognostic marker for MB, corroborating to the role of YAP1 in restricting MB cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(Issue 6):737–754.
Martirosian V, Chen TC, Lin M, Neman J. Medulloblastoma initiation and spread: where neurodevelopment, microenvironment and cancer cross pathways. J Neurosci Res. 2016;94:1511–9.
Fernandez-L A, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–41.
Fernandez-L A, Squatrito M, Northcott P, Awan A, Holland EC, Taylor MD, et al. Oncogenic YAP promotes radioresistance and genomic instability inmedulloblastoma through IGF2-mediated Akt activation. Oncogene. 2012;31:1923–37.
Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74.
Schwalbe EC, Lindsey JC, Nakjang S, Crosier S, Smith AJ, Hicks D, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohortstudy. Lancet Oncol. 2017;18:958–71.
Juan WC, Hong W. Targeting the hippo signaling pathway for tissue regeneration and cancer therapy. Genes. 2016;7:pii: E55.
Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M, et al. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014;588:2663–70.
Imajo M, Ebisuya M, Nishida E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol. 2015;17:7–19.
Ma X, Wang H, Ji J, Xu W, Sun Y, Li W, et al. Hippo signaling promotes JNK-dependent cell migration. Proc Natl Acad Sci USA. 2017;114:1934–9.
Huggett MT, Jermyn M, Gillams A, Illing R, Mosse S, Novelli M. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br J Cancer. 2014;110:1698–704.
Wang LH, Tsai HC, Cheng YC, Lin CY, Huang YL, Tsai CH, et al. CTGF promotes osteosarcoma angiogenesis by regulating miR-543/angiopoietin 2 signaling. Cancer Lett. 2017;391:28–37.
Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71.
Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–93.
Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–40.
Cairns L, Tran T, Kavran JM. Structural insights into the regulation of hippo signaling. ACS Chem Biol. 2017;12:601–10.
Chen L, Loh PG, Song H. Structural and functional insights into the TEAD-YAP complex in the Hippo signaling pathway. Protein Cell. 2010;1:1073–1083.
Mesrouze Y, Bokhovchuk F, Meyerhofer M, Fontana P, Zimmermann C, Martin T, et al. Dissection of the interaction between the intrinsically disordered YAP protein and the transcription fator TEAD. Elife. 2017;6:e25068.
Jóźwiak J, Sontowska I, Bikowska B, Grajkowska W, Galus R, Roszkowski M, et al. Favourable prognosis in medulloblastoma with extensive nodularity is associatedwith mitogen-activated protein kinase upregulation. Folia Neuropathol. 2011;49:257–61.
Korshunov A, Sahm F, Stichel D, Schrimpf D, Ryzhova M, Zheludkova O, et al. Molecularcharacterization of medulloblastomas with extensive nodularity (MBEN). Acta Neuropathol. 2018;136:303–13.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Kumar KS, Pillong M, Kunze J, Burghardt I, Weller M, Grotzer MA, et al. Corrigendum: computer-assisted quantification of motile and invasive capabilities of cancer cells. Sci Rep. 2018;8:46996.
Cruzeiro GAV, Salomão KB, de Biagi CAO Jr, Baumgartner M, Sturm D, Lira RCP, et al. A simplified approach using Taqman low-density array for medulloblastoma subgrouping. Acta Neuropathol Commun. 2019;7:33.
Garzia L, Kijima N, Morrissy AS, De Antonellis P, Guerreiro-Stucklin A, Holgado BL, et al. A hematogenous route for medulloblastoma leptomeningeal metastases. Cell. 2018;172:1050–1062.
Funding
This work was supported by FAPESP grant number 2017/06511–8, 2014/19976–0 and 2014/20341–0.
Author information
Authors and Affiliations
Contributions
G.A.V.C. planned and conducted all experiments, performed the in silico analysis, drafted, and critically read the manuscript. R.C.P.L conducted the overall survival analysis, helped to design the study, and critically read the manuscript. T.A.M., helped to review the design of the study, performed the in silico analysis, and critically read the manuscript. C.A.S. critically read the manuscript, M.B. critically read the manuscript and helped to design the study. L.G.T. provided patient samples and critically read the manuscript. E.T.V. critically read the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This research was submitted to and approved by the HC/FMRP-USP Research Ethics Committee (CAAE no 37206114.1.0000.5440) no 15509/2016.
Informed consent
All samples were obtained after receiving informed consent from all participants included in the study.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cruzeiro, G.A.V., Lira, R.C.P., de Almeida Magalhães, T. et al. CTGF expression is indicative of better survival rates in patients with medulloblastoma. Cancer Gene Ther 27, 378–382 (2020). https://doi.org/10.1038/s41417-019-0100-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0100-3