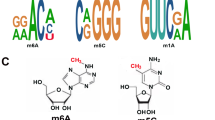
Abstract
Currently, more than 170 modifications have been identified on RNA. Among these RNA modifications, various methylations account for two-thirds of total cases and exist on almost all RNAs. Roles of RNA modifications in cancer are garnering increasing interest. The research on m6A RNA methylation in cancer is in full swing at present. However, there are still many other popular RNA modifications involved in the regulation of gene expression post-transcriptionally besides m6A RNA methylation. In this review, we focus on several important RNA modifications including m1A, m5C, m7G, 2′-O-Me, Ψ and A-to-I editing in cancer, which will provide a new perspective on tumourigenesis by peeking into the complex regulatory network of epigenetic RNA modifications, transcript processing, and protein translation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Not applicable.
References
Yang M, Mo Y, Ren D, Liu S, Zeng Z, Xiong W. Transfer RNA-derived small RNAs in tumor microenvironment. Mol Cancer. 2023;22:32.
Wang J, Ge J, Wang Y, Xiong F, Guo J, Jiang X, et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat Commun. 2022;13:866.
Xiong F, Zhu K, Deng S, Huang H, Yang L, Gong Z, et al. AFAP1-AS1: a rising star among oncogenic long non-coding RNAs. Sci China Life Sci. 2021;64:1602–11.
Wang Y, Yan Q, Mo Y, Liu Y, Wang Y, Zhang S, et al. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumorigenesis via a SERPINH1/c-Myc positive feedback loop. Mol Cancer. 2022;21:62.
Zhao M, Wang Y, Tan F, Liu L, Hou X, Fan C, et al. Circular RNA circCCNB1 inhibits the migration and invasion of nasopharyngeal carcinoma through binding and stabilizing TJP1 mRNA. Sci China Life Sci. 2022;65:2233–47.
Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, et al. Epstein-Barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–88.
Mo Y, Wang Y, Wang Y, Deng X, Yan Q, Fan C, et al. Circular RNA circPVT1 promotes nasopharyngeal carcinoma metastasis via the β-TrCP/c-Myc/SRSF1 positive feedback loop. Mol Cancer. 2022;21:192.
Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231–d5.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200.
Ontiveros RJ, Stoute J, Liu KF. The chemical diversity of RNA modifications. Biochem J. 2019;476:1227–45.
Chen K, Song B, Tang Y, Wei Z, Xu Q, Su J, et al. RMDisease: a database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021;49:D1396–d1404.
Luo X, Li H, Liang J, Zhao Q, Xie Y, Ren J, et al. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021;49:D1405–d12.
Zhi Y, Zhang S, Zi M, Wang Y, Liu Y, Zhang M, et al. Potential applications of N(6) -methyladenosine modification in the prognosis and treatment of cancers via modulating apoptosis, autophagy, and ferroptosis. Wiley Interdiscip Rev RNA. 2022;13:e1719.
Tan F, Zhao M, Xiong F, Wang Y, Zhang S, Gong Z, et al. N6-methyladenosine-dependent signalling in cancer progression and insights into cancer therapies. J Exp Clin Cancer Res. 2021;40:146.
Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198–200.
Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311–6.
Seo KW, Kleiner RE. YTHDF2 recognition of N(1)-methyladenosine (m(1)A)-modified RNA is associated with transcript destabilization. ACS Chem Biol. 2020;15:132–9.
Zheng Q, Gan H, Yang F, Yao Y, Hao F, Hong L, et al. Cytoplasmic m(1)A reader YTHDF3 inhibits trophoblast invasion by downregulation of m(1)A-methylated IGF1R. Cell Discov. 2020;6:12.
Kuang W, Jin H, Yang F, Chen X, Liu J, Li T, et al. ALKBH3-dependent m(1)A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022;8:25.
Woo HH, Chambers SK. Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech. 2019;1862:35–46.
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–6.
Xiong X, Li X, Yi C. N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol. 2018;45:179–86.
Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:816–.e816.
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–45.
Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12:6314.
Richter U, Evans ME, Clark WC, Marttinen P, Shoubridge EA, Suomalainen A, et al. RNA modification landscape of the human mitochondrial tRNA(Lys) regulates protein synthesis. Nat Commun. 2018;9:3966.
Helm M, Giegé R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–46.
Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, et al. Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 2016;14:e1002557.
Sharma S, Hartmann JD, Watzinger P, Klepper A, Peifer C, Kötter P, et al. A single N(1)-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci Rep. 2018;8:11904.
Waku T, Nakajima Y, Yokoyama W, Nomura N, Kako K, Kobayashi A, et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J Cell Sci. 2016;129:2382–93.
Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33.
Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–8.
Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–68.
Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–25.
Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1.
Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90.
Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–.e1111.
Su J, Wu G, Ye Y, Zhang J, Zeng L, Huang X, et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene. 2021;40:5814–28.
Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49:1006–22.
Chen H, Yang H, Zhu X, Yadav T, Ouyang J, Truesdell SS, et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat Commun. 2020;11:2834.
Yang H, Wang Y, Xiang Y, Yadav T, Ouyang J, Phoon L, et al. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc Natl Acad Sci USA. 2022;119:e2116251119.
Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18.
Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47:2041–55.
Shanmugam R, Fierer J, Kaiser S, Helm M, Jurkowski TP, Jeltsch A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015;1:15010.
Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350–62.
Sharma S, Yang J, Watzinger P, Kötter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–76.
Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110.
Dai X, Gonzalez G, Li L, Li J, You C, Miao W, et al. YTHDF2 binds to 5-methylcytosine in RNA and modulates the maturation of ribosomal RNA. Anal Chem. 2020;92:1346–54.
Heissenberger C, Liendl L, Nagelreiter F, Gonskikh Y, Yang G, Stelzer EM, et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019;47:11807–25.
Boulias K, Greer EL. Put the pedal to the METTL1: adding internal m(7)G increases mRNA translation efficiency and augments miRNA processing. Mol Cell. 2019;74:1105–7.
Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–7.
Ohira T, Suzuki T. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat Chem Biol. 2016;12:648–55.
Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G, et al. Transcriptome-wide mapping of internal N(7)-Methylguanosine methylome in mammalian mRNA. Mol Cell. 2019;74:1304–.e1308.
Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 2019;29:927–41.
Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci. 2018;19:4080.
Zhu C, Yan Q, Weng C, Hou X, Mao H, Liu D, et al. Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc Natl Acad Sci USA. 2018;115:10082–7.
Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N, et al. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol Cell. 2019;74:1278–.e1279.
Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W, et al. N(7)-methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339–.e3338.
Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci USA. 2016;113:5263–8.
Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–51.
Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244–.e245.
Adamiak DA, Milecki J, Popenda M, Adamiak RW, Dauter Z, Rypniewski WR. Crystal structure of 2′-O-Me(CGCGCG)2, an RNA duplex at 1.30 A resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res. 1997;25:4599–607.
Lacoux C, Di Marino D, Boyl PP, Zalfa F, Yan B, Ciotti MT, et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012;40:4086–96.
Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, et al. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc Natl Acad Sci USA. 2017;114:E2106–e2115.
Elliott BA, Ho HT, Ranganathan SV, Vangaveti S, Ilkayeva O, Abou Assi H, et al. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat Commun. 2019;10:3401.
Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. Rna. 2009;15:675–85.
Picard-Jean F, Brand C, Tremblay-Létourneau M, Allaire A, Beaudoin MC, Boudreault S, et al. 2′-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS ONE. 2018;13:e0193804.
Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods. 2017;14:695–8.
Choi J, Indrisiunaite G, DeMirci H, Ieong KW, Wang J, Petrov A, et al. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat Struct Mol Biol. 2018;25:208–16.
Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochim Biophys Acta Gene Regul Mech. 2019;1862:253–69.
Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–51.
Monaco PL, Marcel V, Diaz JJ, Catez F. 2′-O-methylation of ribosomal RNA: towards an epitranscriptomic control of translation? Biomolecules. 2018;8:106.
Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci USA. 2002;99:12697–702.
Lin TY, Mehta R, Glatt S. Pseudouridines in RNAs-switching atoms means shifting paradigms. FEBS Lett. 2021;595:2310–22.
Malik TN, Doherty EE, Gaded VM, Hill TM, Beal PA, Emeson RB. Regulation of RNA editing by intracellular acidification. Nucleic Acids Res. 2021;49:4020–36.
Shen P, Yang T, Chen Q, Yuan H, Wu P, Cai B, et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol Cancer. 2021;20:51.
Zhang L, Yang CS, Varelas X, Monti S. Altered RNA editing in 3′ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci Rep. 2016;6:23226.
Ma CP, Liu H, Yi-Feng Chang I, Wang WC, Chen YT, Wu SM, et al. ADAR1 promotes robust hypoxia signaling via distinct regulation of multiple HIF-1α-inhibiting factors. EMBO Rep. 2019;20:e47107.
Jiang L, Hao Y, Shao C, Wu Q, Prager BC, Gimple RC, et al. ADAR1-mediated RNA editing links ganglioside catabolism to glioblastoma stem cell maintenance. J Clin Invest. 2022;132:e143397.
Fu L, Qin YR, Ming XY, Zuo XB, Diao YW, Zhang LY, et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci USA. 2017;114:E4631–e4640.
Brümmer A, Yang Y, Chan TW, Xiao X. Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat Commun. 2017;8:1255.
Nakano M, Fukami T, Gotoh S, Nakajima M. A-to-I RNA editing up-regulates human dihydrofolate reductase in breast cancer. J Biol Chem. 2017;292:4873–84.
Ramírez-Moya J, Miliotis C, Baker AR, Gregory RI, Slack FJ, Santisteban P. An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks. Mol Cancer. 2021;20:115.
Han F, Hu M, Zhang L, Fan X, Wang J, Lou Z, et al. A-to-I RNA editing of BLCAP promotes cell proliferation by losing the inhibitory of Rb1 in colorectal cancer. Exp Cell Res. 2022;417:113209.
Song Y, An O, Ren X, Chan THM, Tay DJT, Tang SJ, et al. RNA editing mediates the functional switch of COPA in a novel mechanism of hepatocarcinogenesis. J Hepatol. 2021;74:135–47.
Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–16.
Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Kim KY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med. 2014;211:613–21.
Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17:311–21.
Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell. 2016;19:177–91.
Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21.
Wang Y, Xu X, Yu S, Jeong KJ, Zhou Z, Han L, et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017;27:1112–25.
Ramírez-Moya J, Baker AR, Slack FJ, Santisteban P. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene. 2020;39:3738–53.
Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, et al. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 2018;46:2045–59.
Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122:4059–76.
Paul D, Sinha AN, Ray A, Lal M, Nayak S, Sharma A, et al. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci Rep. 2017;7:2466.
Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. Rna. 2005;11:1281–90.
Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017;68:993–1005.e1009.
Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R, et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun. 2022;13:2165.
Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. Rna. 2012;18:2269–76.
Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251–5.
Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–93.
Oerum S, Roovers M, Rambo RP, Kopec J, Bailey HJ, Fitzpatrick F, et al. Structural insight into the human mitochondrial tRNA purine N1-methyltransferase and ribonuclease P complexes. J Biol Chem. 2018;293:12862–76.
Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401–15.
Wagner A, Hofmeister O, Rolland SG, Maiser A, Aasumets K, Schmitt S, et al. Mitochondrial Alkbh1 localizes to mtRNA granules and its knockdown induces the mitochondrial UPR in humans and C. elegans. J Cell Sci. 2019;132:jcs223891.
Rashad S, Han X, Sato K, Mishima E, Abe T, Tominaga T, et al. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020;17:1092–103.
Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–63.
Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, et al. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–16.
Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–.e975.
Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90:6380–4.
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8.
Xue S, Xu H, Sun Z, Shen H, Chen S, Ouyang J, et al. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration. Biochem Biophys Res Commun. 2019;520:60–66.
Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006;34:6034–43.
Sajini AA, Choudhury NR, Wagner RE, Bornelöv S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10:2550.
Henry BA, Kanarek JP, Kotter A, Helm M, Lee N. 5-methylcytosine modification of an Epstein-Barr virus noncoding RNA decreases its stability. Rna. 2020;26:1038–48.
Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34:3630–41.
Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546–51.
Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039.
Yang L, Ren Z, Yan S, Zhao L, Liu J, Zhao L, et al. Nsun4 and Mettl3 mediated translational reprogramming of Sox9 promotes BMSC chondrogenic differentiation. Commun Biol. 2022;5:495.
Haag S, Warda AS, Kretschmer J, Günnigmann MA, Höbartner C, Bohnsack MT. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. Rna. 2015;21:1532–43.
Liu J, Huang T, Zhang Y, Zhao T, Zhao X, Chen W, et al. Sequence- and structure-selective mRNA m(5)C methylation by NSUN6 in animals. Natl Sci Rev. 2021;8:nwaa273.
Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 2016;14:479–92.
Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem. 2015;16:752–5.
Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM, et al. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J Biol Chem. 2021;296:100087.
Zhang Y, Wang C. Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases. Mol Biol Rep. 2021;48:4747–56.
Eckwahl M, Xu R, Michalkiewicz J, Zhang W, Patel P, Cai Z, et al. 5-Methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J Virol. 2020;94:e00544–20.
Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC, et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. 2021;41:560–75.
Zheng H, Zhu M, Li W, Zhou Z, Wan X. m(5) C and m(6) A modification of long noncoding NKILA accelerates cholangiocarcinoma progression via the miR-582-3p-YAP1 axis. Liver Int. 2022;42:1144–57.
Gao W, Chen L, Lin L, Yang M, Li T, Wei H, et al. SIAH1 reverses chemoresistance in epithelial ovarian cancer via ubiquitination of YBX-1. Oncogenesis. 2022;11:13.
Yan J, Liu J, Huang Z, Huang W, Lv J. FOXC2-AS1 stabilizes FOXC2 mRNA via association with NSUN2 in gastric cancer cells. Hum Cell. 2021;34:1755–64.
Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–74.
Trotman JB, Giltmier AJ, Mukherjee C, Schoenberg DR. RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res. 2017;45:10726–39.
Gonatopoulos-Pournatzis T, Dunn S, Bounds R, Cowling VH. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell. 2011;44:585–96.
Bueren-Calabuig JA, G Bage M, Cowling VH, Pisliakov AV. Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations. Nucleic Acids Res. 2019;47:8675–92.
Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. Rna. 2002;8:1253–66.
Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T, et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422–35.
Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu H, et al. METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin Transl Med. 2021;11:e661.
Chen J, Li K, Chen J, Wang X, Ling R, Cheng M, et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun. 2022;42:223–44.
Chen B, Jiang W, Huang Y, Zhang J, Yu P, Wu L, et al. N(7)-methylguanosine tRNA modification promotes tumorigenesis and chemoresistance through WNT/β-catenin pathway in nasopharyngeal carcinoma. Oncogene. 2022;41:2239–53.
Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X, et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. 2021;11:e675.
Zhao Y, Kong L, Pei Z, Li F, Li C, Sun X, et al. m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front Cell Dev Biol. 2021;9:642080.
Létoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, et al. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci USA. 2014;111:E5518–5526.
Haag S, Kretschmer J, Bohnsack MT. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. Rna. 2015;21:180–7.
Joshi B, Lee K, Maeder DL, Jagus R. Phylogenetic analysis of eIF4E-family members. BMC Evolut Biol. 2005;5:48.
Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33:227–38.
Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6:65–69.
Zhang X, Chapat C, Wang P, Choi JH, Li Q, Luo J, et al. microRNA-induced translational control of antiviral immunity by the cap-binding protein 4EHP. Mol Cell. 2021;81:1187–.e1185.
Christie M, Igreja C. eIF4E-homologous protein (4EHP): a multifarious cap-binding protein. FEBS J. 2023;290;266–85.
Jeong SJ, Park S, Nguyen LT, Hwang J, Lee EY, Giong HK, et al. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat Commun. 2019;10:1357.
von Stechow L, Typas D, Carreras Puigvert J, Oort L, Siddappa R, Pines A, et al. The E3 ubiquitin ligase ARIH1 protects against genotoxic stress by initiating a 4EHP-mediated mRNA translation arrest. Mol Cell Biol. 2015;35:1254–68.
Volpon L, Osborne MJ, Culjkovic-Kraljacic B, Borden KL. eIF4E3, a new actor in mRNA metabolism and tumor suppression. Cell Cycle. 2013;12:1159–60.
Weiss B, Allen GE, Kloehn J, Abid K, Jaquier-Gubler P, Curran JA. eIF4E3 forms an active eIF4F complex during stresses (eIF4FS) targeting mTOR and re-programs the translatome. Nucleic Acids Res. 2021;49:5159–76.
Li H, Dong H, Xu B, Xiong QP, Li CT, Yang WQ, et al. A dual role of human tRNA methyltransferase hTrmt13 in regulating translation and transcription. EMBOJ. 2022;41:e108544.
Guy MP, Phizicky EM. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. Rna. 2015;21:61–74.
Nagayoshi Y, Chujo T, Hirata S, Nakatsuka H, Chen CW, Takakura M, et al. Loss of Ftsj1 perturbs codon-specific translation efficiency in the brain and is associated with X-linked intellectual disability. Sci Adv. 2021;7:eabf3072.
Li J, Wang YN, Xu BS, Liu YP, Zhou M, Long T, et al. Intellectual disability-associated gene ftsj1 is responsible for 2′-O-methylation of specific tRNAs. EMBO Rep. 2020;21:e50095.
Freund I, Buhl DK, Boutin S, Kotter A, Pichot F, Marchand V, et al. 2′-O-methylation within prokaryotic and eukaryotic tRNA inhibits innate immune activation by endosomal Toll-like receptors but does not affect recognition of whole organisms. Rna. 2019;25:869–80.
Lee KW, Bogenhagen DF. Assignment of 2′-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16 S ribosomal RNA (rRNA). J Biol Chem. 2014;289:24936–42.
Simabuco FM, Morello LG, Aragão AZ, Paes Leme AF, Zanchin NI. Proteomic characterization of the human FTSJ3 preribosomal complexes. J Proteome Res. 2012;11:3112–26.
Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019;565:500–4.
Liang H, Jiao Z, Rong W, Qu S, Liao Z, Sun X, et al. 3′-Terminal 2′-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020;48:7027–40.
Lim SL, Qu ZP, Kortschak RD, Lawrence DM, Geoghegan J, Hempfling AL, et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015;11:e1005620.
Bélanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem. 2010;285:33037–44.
Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, et al. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011;39:4756–68.
Rintala-Dempsey AC, Kothe U. Eukaryotic stand-alone pseudouridine synthases—RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185–96.
Borchardt EK, Martinez NM, Gilbert WV. Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet. 2020;54:309–36.
Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B, et al. mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol. 2019;15:966–74.
Martinez NM, Su A, Burns MC, Nussbacher JK, Schaening C, Sathe S, et al. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol Cell. 2022;82:645–.e649.
Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135:707–13.
Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27:393–406.
Mukhopadhyay S, Deogharia M, Gupta R. Mammalian nuclear TRUB1, mitochondrial TRUB2, and cytoplasmic PUS10 produce conserved pseudouridine 55 in different sets of tRNA. Rna. 2021;27:66–79.
Antonicka H, Choquet K, Lin ZY, Gingras AC, Kleinman CL, Shoubridge EA. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 2017;18:28–38.
Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–.e1226.
Zhang DY, Ming GL, Song H. PUS7: a targetable epitranscriptomic regulator of glioblastoma growth. Trends Pharmacol Sci. 2021;42:976–8.
Zaganelli S, Rebelo-Guiomar P, Maundrell K, Rozanska A, Pierredon S, Powell CA, et al. The Pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules. J Biol Chem. 2017;292:4519–32.
Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020;16:160–9.
Kamalampeta R, Kothe U. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep. 2012;2:663.
Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell. 2009;34:427–39.
Caton EA, Kelly EK, Kamalampeta R, Kothe U. Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 2018;46:905–16.
Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma 2005;114:1–14.
Levi O, Arava YS. Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 2021;49:432–43.
Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBOJ. 2016;35:654–67.
Schaub M, Keller W. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie. 2002;84:791–803.
Eisenberg E, Levanon EY. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet. 2018;19:473–90.
Keegan LP, Leroy A, Sproul D, OConnell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209.
Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–8.
Oakes E, Anderson A, Cohen-Gadol A, Hundley HA. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma. J Biol Chem. 2017;292:4326–35.
Torres AG, Piñeyro D, Filonava L, Stracker TH, Batlle E, Ribas de Pouplana L. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 2014;588:4279–86.
Zheng Q, Yu X, Zhang Q, He Y, Guo W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci Rep. 2021;41:BSR20210337.
Gao Y, Wang H, Li H, Ye X, Xia Y, Yuan S, et al. Integrated analyses of m(1)A regulator-mediated modification patterns in tumor microenvironment-infiltrating immune cells in colon cancer. Oncoimmunology. 2021;10:1936758.
Liu J, Chen C, Wang Y, Qian C, Wei J, Xing Y, et al. Comprehensive of N1-methyladenosine modifications patterns and immunological characteristics in ovarian cancer. Front Immunol. 2021;12:746647.
Gao L, Chen R, Sugimoto M, Mizuta M, Kishimoto Y, Omori K. The impact of m1A methylation modification patterns on tumor immune microenvironment and prognosis in oral squamous cell carcinoma. Int J Mol Sci. 2021;22:19.
Esteve-Puig R, Climent F, Piñeyro D, Domingo-Domènech E, Davalos V, Encuentra M, et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood. 2021;137:994–9.
Zhao Y, Zhao Q, Kaboli PJ, Shen J, Li M, Wu X, et al. m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer. Transl Oncol. 2019;12:1323–33.
Shi L, Yang XM, Tang DD, Liu G, Yuan P, Yang Y, et al. Expression and significance of m1A transmethylase, hTrm6p/hTrm61p and its related gene hTrm6/hTrm61 in bladder urothelial carcinoma. Am J Cancer Res. 2015;5:2169–79.
Zhang Q, Zheng Q, Yu X, He Y, Guo W. Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J Transl Med. 2020;18:245.
Huang W, Qi CB, Lv SW, Xie M, Feng YQ, Huang WH, et al. Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal Chem. 2016;88:1378–84.
Wang L, Zhang J, Su Y, Maimaitiyiming Y, Yang S, Shen Z, et al. Distinct roles of m(5)C RNA methyltransferase NSUN2 in major gynecologic cancers. Front Oncol. 2022;12:786266.
Chen L, Ding J, Wang B, Chen X, Ying X, Yu Z, et al. RNA methyltransferase NSUN2 promotes hypopharyngeal squamous cell carcinoma proliferation and migration by enhancing TEAD1 expression in an m(5)C-dependent manner. Exp Cell Res. 2021;404:112664.
Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842.
Liu J, Ren Z, Yang L, Zhu L, Li Y, Bie C, et al. The NSUN5-FTH1/FTL pathway mediates ferroptosis in bone marrow-derived mesenchymal stem cells. Cell Death Discov. 2022;8:99.
Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020;11:270.
Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906–19.
Piserà A, Campo A, Campo S. Structure and functions of the translation initiation factor eIF4E and its role in cancer development and treatment. J Genet Genomics = Yi Chuan Xue Bao. 2018;45:13–24.
Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene 2010;29:930–6.
Posternak V, Ung MH, Cheng C, Cole MD. MYC mediates mRNA cap methylation of canonical Wnt/β-catenin signaling transcripts by recruiting CDK7 and RNA methyltransferase. Mol Cancer Res. 2017;15:213–24.
Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R, et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022;13:1478.
Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W, et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323–.e3314.
Liu Y, Yang C, Zhao Y, Chi Q, Wang Z, Sun B. Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging. 2019;11:12328–44.
Jansson MD, Häfner SJ, Altinel K, Tehler D, Krogh N, Jakobsen E, et al. Regulation of translation by site-specific ribosomal RNA methylation. Nat Struct Mol Biol. 2021;28:889–99.
Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Köhn M, et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol. 2017;19:844–55.
Pauli C, Liu Y, Rohde C, Cui C, Fijalkowska D, Gerloff D, et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood. 2020;135:2059–70.
Metge BJ, Kammerud SC, Pruitt HC, Shevde LA, Samant RS. Hypoxia re-programs 2′-O-Me modifications on ribosomal RNA. iScience. 2021;24:102010.
Yi Y, Li Y, Meng Q, Li Q, Li F, Lu B, et al. A PRC2-independent function for EZH2 in regulating rRNA 2′-O methylation and IRES-dependent translation. Nat Cell Biol. 2021;23:341–54.
Guerrieri AN, Zacchini F, Onofrillo C, Di Viggiano S, Penzo M, Ansuini A, et al. DKC1 overexpression induces a more aggressive cellular behavior and increases intrinsic ribosomal activity in immortalized mammary gland cells. Cancers. 2020;12:3512.
Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–35.
Cui Q, Yin K, Zhang X, Ye P, Chen X, Chao J, et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat Cancer. 2021;2:932–49.
Kan G, Wang Z, Sheng C, Chen G, Yao C, Mao Y, et al. Dual inhibition of DKC1 and MEK1/2 synergistically restrains the growth of colorectal cancer cells. Adv Sci. 2021;8:2004344.
Peng X, Xu X, Wang Y, Hawke DH, Yu S, Han L, et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018;33:817–.e817.
Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–28.
Ruan H, Li Q, Liu Y, Liu Y, Lussier C, Diao L, et al. GPEdit: the genetic and pharmacogenomic landscape of A-to-I RNA editing in cancers. Nucleic Acids Res. 2022;50:D1231–d1237.
Galeano F, Rossetti C, Tomaselli S, Cifaldi L, Lezzerini M, Pezzullo M, et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32:998–1009.
Chen YB, Liao XY, Zhang JB, Wang F, Qin HD, Zhang L, et al. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int J Oncol. 2017;50:622–30.
Gumireddy K, Li A, Kossenkov AV, Sakurai M, Yan J, Li Y, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat Commun. 2016;7:10715.
Peng L, Zhang H, Su Y, Shen Q, Du C, Xie H, et al. Lipopolysaccharide enhances ADAR2 which drives Hirschsprung′s disease by impairing miR-142-3p biogenesis. J Cell Mol Med. 2018;22:4045–55.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Jiang X, Deng X, Wang J, Mo Y, Shi L, Wei F, et al. BPIFB1 inhibits vasculogenic mimicry via downregulation of GLUT1-mediated H3K27 acetylation in nasopharyngeal carcinoma. Oncogene. 2022;41:233–45.
Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20:112.
Zhong Y, Yang L, Xiong F, He Y, Tang Y, Shi L, et al. Long non-coding RNA AFAP1-AS1 accelerates lung cancer cells migration and invasion by interacting with SNIP1 to upregulate c-Myc. Signal Transduct Target Ther. 2021;6:240.
Fan C, Zhang S, Gong Z, Li X, Xiang B, Deng H, et al. Emerging role of metabolic reprogramming in tumor immune evasion and immunotherapy. Sci China Life Sci. 2021;64:534–47.
Tang L, Xiong W, Zhang L, Wang D, Wang Y, Wu Y, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene. 2021;40:307–21.
Fan C, Qu H, Xiong F, Tang Y, Tang T, Zhang L, et al. CircARHGAP12 promotes nasopharyngeal carcinoma migration and invasion via ezrin-mediated cytoskeletal remodeling. Cancer Lett. 2021;496:41–56.
Wang M, Dai M, Wang D, Tang T, Xiong F, Xiang B, et al. The long noncoding RNA AATBC promotes breast cancer migration and invasion by interacting with YBX1 and activating the YAP1/Hippo signaling pathway. Cancer Lett. 2021;512:60–72.
Wei X, Chen Y, Jiang X, Peng M, Liu Y, Mo Y, et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol Cancer. 2021;20:7.
Dai DL, Li X, Wang L, Xie C, Jin Y, Zeng MS, et al. Identification of an N6-methyladenosine-mediated positive feedback loop that promotes Epstein-Barr virus infection. J Biol Chem. 2021;296:100547.
Chi Y, Liang Z, Guo Y, Chen D, Lu L, Lin J, et al. WBSCR22 confers cell survival and predicts poor prognosis in glioma. Brain Res Bull. 2020;161:1–12.
Khan AA, Huang H, Zhao Y, Li H, Pan R, Wang S, et al. WBSCR22 and TRMT112 synergistically suppress cell proliferation, invasion and tumorigenesis in pancreatic cancer via transcriptional regulation of ISG15. Int J Oncol. 2022;60:24.
Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–6.
Nachmani D, Bothmer AH, Grisendi S, Mele A, Bothmer D, Lee JD, et al. Germline NPM1 mutations lead to altered rRNA 2′-O-methylation and cause dyskeratosis congenita. Nat Genet. 2019;51:1518–29.
Wang B, Niu L, Wang Z, Zhao Z. RNA m1A methyltransferase TRMT6 predicts poorer prognosis and promotes malignant behavior in glioma. Front Mol Biosci. 2021;8:692130.
Wang Q, Zhang Q, Huang Y, Zhang J. m(1)A regulator TRMT10C predicts poorer survival and contributes to malignant behavior in gynecological cancers. DNA Cell Biol. 2020;39:1767–78.
Jiang Z, Li S, Han MJ, Hu GM, Cheng P. High expression of NSUN5 promotes cell proliferation via cell cycle regulation in colorectal cancer. Am J Transl Res. 2020;12:3858–70.
Liu Y, Zhang Y, Chi Q, Wang Z, Sun B. Methyltransferase-like 1 (METTL1) served as a tumor suppressor in colon cancer by activating 7-methyguanosine (m7G) regulated let-7e miRNA/HMGA2 axis. Life Sci. 2020;249:117480.
Tian QH, Zhang MF, Zeng JS, Luo RG, Wen Y, Chen J, et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J Mol Med. 2019;97:1535–45.
Wang C, Wang W, Han X, Du L, Li A, Huang G. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol Lett. 2021;21:330.
Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang G, et al. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 2021;12:691.
Yan D, Tu L, Yuan H, Fang J, Cheng L, Zheng X, et al. WBSCR22 confers oxaliplatin resistance in human colorectal cancer. Sci Rep. 2017;7:15443.
Zhao H, Su W, Sun Y, Wu Z. WBSCR22 competes with long non-coding RNA Linc00346 for miR-509-5p binding site to regulate cancer stem cell phenotypes of colorectal cancer. Biochem Genet. 2020;58:384–98.
Zhao H, Su W, Kang Q, Xing Z, Lin X, Wu Z. Natural killer cells inhibit oxaliplatin-resistant colorectal cancer by repressing WBSCR22 via upregulating microRNA-146b-5p. Am J Cancer Res. 2018;8:824–34.
Hou P, Shi P, Jiang T, Yin H, Chu S, Shi M, et al. DKC1 enhances angiogenesis by promoting HIF-1α transcription and facilitates metastasis in colorectal cancer. Br J Cancer. 2020;122:668–79.
Kan G, Wang Z, Sheng C, Yao C, Mao Y, Chen S. Inhibition of DKC1 induces telomere-related senescence and apoptosis in lung adenocarcinoma. J Transl Med. 2021;19:161.
Zhang M, Pan Y, Jiang R, Hou P, Shan H, Chen F, et al. DKC1 serves as a potential prognostic biomarker for human clear cell renal cell carcinoma and promotes its proliferation, migration and invasion via the NF‑κB pathway. Oncol Rep. 2018;40:968–78.
O’Brien R, Tran SL, Maritz MF, Liu B, Kong CF, Purgato S, et al. MYC-driven neuroblastomas are addicted to a telomerase-independent function of dyskerin. Cancer Res. 2016;76:3604–17.
Song D, Guo M, Xu S, Song X, Bai B, Li Z, et al. HSP90-dependent PUS7 overexpression facilitates the metastasis of colorectal cancer cells by regulating LASP1 abundance. J Exp Clin Cancer Res: CR. 2021;40:170.
Du J, Gong A, Zhao X, Wang G. Pseudouridylate synthase 7 promotes cell proliferation and invasion in colon cancer through activating PI3K/AKT/mTOR signaling pathway. Dig Dis Sci. 2022;67:1260–70.
He Q, Yang L, Gao K, Ding P, Chen Q, Xiong J, et al. FTSJ1 regulates tRNA 2′-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 2020;11:348.
Liu B, Li J, Zheng M, Ge J, Li J, Yu P. MiR-542-3p exerts tumor suppressive functions in non-small cell lung cancer cells by upregulating FTSJ2. Life Sci. 2017;188:87–95.
Manning M, Jiang Y, Wang R, Liu L, Rode S, Bonahoom M, et al. Pan-cancer analysis of RNA methyltransferases identifies FTSJ3 as a potential regulator of breast cancer progression. RNA Biol. 2020;17:474–86.
Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95.
Velazquez-Torres G, Shoshan E, Ivan C, Huang L, Fuentes-Mattei E, Paret H, et al. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun. 2018;9:461.
Kung CP, Cottrell KA, Ryu S, Bramel ER, Kladney RD, Bao EA, et al. Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer. Oncogene. 2021;40:189–202.
Liu X, Fu Y, Huang J, Wu M, Zhang Z, Xu R, et al. ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation. J Exp Clin Cancer Res. 2019;38:315.
Zhang Y, Wang K, Zhao Z, Sun S, Zhang K, Huang R, et al. ADAR3 expression is an independent prognostic factor in lower-grade diffuse gliomas and positively correlated with the editing level of GRIA2(Q607R). Cancer Cell Int. 2018;18:196.
Acknowledgements
The figures are created with Biorender.com.
Funding
This study was supported by grants from the National Natural Science Foundation of China (U21A20382, U20A20367 and 82072374), the Overseas Expertise Introduction Project for Discipline Innovation (BP1221008), the Natural Science Foundation of Hunan Province (2021JJ30897).
Author information
Authors and Affiliations
Contributions
TQL, LLY, WYM, WP, HXC, OYJW, FCM, LZ, WFY, GC, ZM, LQJ and WH collected the relevant literature and drafted the manuscript. XB, JWH, LGY, ZZY and XW participated in the inception and revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agreed to publication.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, Q., Li, L., Wang, Y. et al. RNA modifications in cancer. Br J Cancer 129, 204–221 (2023). https://doi.org/10.1038/s41416-023-02275-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02275-1
This article is cited by
-
Antizyme inhibitor family: biological and translational research implications
Cell Communication and Signaling (2024)
-
Epigenetic regulation of diverse cell death modalities in cancer: a focus on pyroptosis, ferroptosis, cuproptosis, and disulfidptosis
Journal of Hematology & Oncology (2024)
-
Research progress of N1-methyladenosine RNA modification in cancer
Cell Communication and Signaling (2024)
-
Digital RNA sequencing using unique molecular identifiers enables ultrasensitive RNA mutation analysis
Communications Biology (2024)
-
METTL3 and METTL14-mediated N6-methyladenosine modification of SREBF2-AS1 facilitates hepatocellular carcinoma progression and sorafenib resistance through DNA demethylation of SREBF2
Scientific Reports (2024)