Abstract
Epidermal growth factor receptor (EGFR) mutations in lung adenocarcinoma are a frequent class of driver mutations. Single EGFR tyrosine kinase inhibitor (TKI) provides substantial clinical benefit, but almost nil radiographic complete responses. Patients invariably progress, although survival can reach several years with post-treatment therapies, including EGFR TKIs, chemotherapy or other procedures. Endeavours have been clinically oriented to manage the acquisition of EGFR TKI-resistant mutations; however, basic principles on cancer evolution have not been considered in clinical trials. For years, evidence has displayed rapidly adaptive mechanisms of resistance to selective monotherapy, posing several dilemmas for the practitioner. Strict adherence to non-small cell lung cancer (NSCLC) guidelines is not always practical for addressing the clinical progression that EGFR-mutant lung adenocarcinoma patients suffer. The purpose of this review is to highlight regulatory mechanisms and signalling pathways that cause therapy-induced resistance to EGFR TKIs. It suggests combinatorial therapies that target EGFR, as well as potential mechanisms underlying EGFR-mutant NSCLC, alerting the reader to clinical opportunities that may lead to a deeper and more durable response. Molecular reprogramming contributes to EGFR TKI resistance, and the compiled information is relevant in understanding the development of new combined targeted strategies in EGFR-mutant NSCLC.
Similar content being viewed by others
Background
Epidermal growth factor receptor (EGFR) mutations, such as in-frame deletion of the amino acids LREA within exon 19 and L858R substitution mutation, activate EGFR, leading to proliferation and anti-apoptotic signalling. Lung adenocarcinoma (LUAD) patients harbouring such EGFR gain-of-function mutations are sensitive to EGFR tyrosine kinase inhibitors (TKIs). LUAD patients benefit from single therapy with EGFR TKI, leading to tumour shrinkage and prolonged progression-free survival (PFS) and overall survival (OS). Notwithstanding, all patients with TKI-sensitising EGFR mutations will recur after the initial response to TKIs. Adaptive resistance to standalone receptor tyrosine kinase (RTK) inhibitors invariably occurs via kinome signalling rewiring in multiple forms of cancer [1]. Loss of the protein tyrosine phosphatase PTPN12 activates diverse RTKs, such as EGFR and HER2, in triple-negative breast cancer [2]. PTPN12 in triple-negative breast cancer acts as a negative regulator of HER2/EGFR and other TKs, such as PDGFR-β and ABL (Abelson murine leukaemia viral oncogene). Lapatinib in combination with sunitinib reduces the proliferation in triple-negative breast cancer cells [2]. The F-box protein β-transducin repeat-containing protein (β-TRCP) negatively regulates the REST tumour suppressor that positively regulates PTPN12 levels, partly by inhibiting miR-124. PTPN12 is deleted (on chromosome 7) in lung cancer (13.8%) and miR-124 is focally amplified (minimal region of amplification in chromosome 20) in breast and lung cancer (20.1% and 12.1%, respectively). Several lung cancer cell lines (i.e. PC9 exon 19 deletion) harbour focal amplification in chromosome 20 (miR-124). Interestingly, the EGFR Y1148 residue showed the strongest differential phosphorylation (>2-fold) in response to PTPN12 depletion [2]. The use of RTK inhibitor combinations to simultaneously target several RTKs has been suggested as a strategy for personalised cancer therapy. Single RTK inhibitors did not induce the activation of new RTKs, but only increased the phosphorylation of the existing activated RTKs to compensate for the reduced phosphorylation of ERK or Akt [3]. A proposed model for alternative activation of RTKs upon activation of mesenchymal-epithelial transition (MET), by its ligand HGF (from stromal cells), induced interreceptor crosstalk with integrin beta-4 (ITGB4), erythropoietin-producing hepatocellular (EphA2), CUB domain-containing protein-1 (CDCP), AXL and Jak1, providing an alternative signalling mechanism and, henceforth, circumventing EGFR TKI effect [4]. We previously found that gefitinib or osimertinib activates STAT3 and Src-YAP1 (Yes-associated protein 1) in EGFR mutation-positive non-small cell lung cancer (NSCLC) cells [5] and that genetic or pharmacologic inhibition of Src or SFKs diminishes YAP1, AXL and CDCP1 phosphorylation or expression. In two cohorts of EGFR mutation-positive NSCLC patients treated with EGFR TKI, we identified that a risk model combining AXL and CDCP1 messenger RNA (mRNA) expression was strongly associated with PFS (hazard ratios of 2.95 and 2.19) and OS (hazard ratios of 3.56 and 2.96) between high- and low-risk group. We postulated that Src-YAP1 signalling leads to further activation of AXL, CDCP1 and MET [6]. Activated AXL has been associated with EGFR and HER3 in maintaining cell survival and inducing drug-tolerant cells to osimertinib [7]. Via other RTKs, parallel bypass pathways have been comprehensively reviewed by Rotow and Bivona [8] and by Tomasello et al. [9]. RTK phosphorylation/activation profiles have been analysed from different tissue origins, including lung cancer. Similar results were obtained from the cancer cell lines, the primary cancer samples and the xenograft samples that had more than one RTK activated [3]. The mechanism of activation of multiple RTKs is multifarious, including phosphatase PTPN12 that serves as a feedback mechanism to limit receptor signalling in triple-negative breast cancer. The suite of RTK substrates inhibited by PTPN12 encompasses MET, PDGFRβ, EGFR and others [2, 10]. New data indicate that targeting AXL expressed on tumour cells and MERTK in the tumour microenvironment is predicted to enhance immunotherapy activity. Tumour-associated M2 macrophages express MERTK and the inhibition of MERTK blocks the immune-suppressive effects of macrophage efferocytosis [11]. Adding further evidence, GAS6/MERTK signalling has been seen to stimulate NSCLC proliferation. Nuclear PD-L1 regulates the synthesis of GAS6 mRNA, promoting GAS6 secretion to activate MERTK signalling pathway [12]. Interestingly, epiregulin (an EGFR ligand) is predominantly expressed in macrophages in the tumour microenvironment. Epiregulin secreted by macrophages induces the formation of the EGFR/ERBB2 heterodimer and causes resistance to EGFR TKI in an EGFR/ERBB2-AKY axis-dependent manner [13].
Assessment of clinical specimens has shown a ceaseless number of resistance mechanisms to either first- or second-generation EGFR TKIs (i.e. erlotinib, afatinib) or third-generation EGFR TKIs (i.e. osimertinib) that have principally included up-regulation or amplification of AXL [7, 14], Her-2, MET, Akt [15], ERK and nuclear factor-κB (NF-κB) signalling [16]. Similarly, EGFR second-site mutation T790M often reappears post osimertinib treatment, while other EGFR mutations are acquired, such as EGFR C797S, G724, L792 and L718/G719 [17, 18]. Variegation of other genetic defects has been found following first-line osimertinib or later-line osimertinib, such as RET fusions, BRAF fusions, K-Ras mutations, MET amplification and MET H1094Y mutations, FGFR amplification and histological transformation to small cell lung cancer and squamous cell carcinoma [17]. Therefore, the physician is confronted with a broad array of resistance mechanisms either pretreatment or post-progression in the analysis of tumour samples and circulating tumour DNA in the plasma. Next-generation sequencing (NGS) platforms, as well as custom RNA-sequencing panels, provide extensive information on each individual EGFR-mutant lung cancer patient. Minion combinatory therapies are being developed and signalling targetable pathways have been charted [19].
PRKCD (protein kinase Cδ) a mediator of TKI resistance
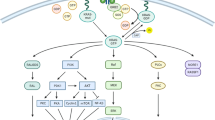
EGFR TKIs, such as gefitinib, induced EGFR heterodimers in TKI-resistant EGFR-mutant NSCLC, where phosphorylation of EGFR Y845 and Y1086 was almost suppressed. However, EGFR pY1173 was only partially reduced by gefitinib and promoted activation of phospholipase γ2 (PLCγ2) and PKCδ when sustained by EGFR heterodimer [20] (Fig. 1a). Phosphatase and tensin homologue (PTEN) loss [21] and Akt and NF-κB pathway activation are present in H1650 cells (EGFR-del19 mutation) resistant to TKIs [16]. Intriguingly, EGFR depletion attenuated all Akt, ERK and RelA phosphorylation compared with EGFR kinase inhibition, which did not affect Akt or RelA. Similar effects of EGFR depletion were noted in two other NSCLC cell lines, H1975 (EGFR L858R/T790M mutation) and H820 (EGFR-del19/T790M mutation and MET amplification) [20]. Of the 15 HCC827 gefitinib-resistant clones (median inhibitory concentration [IC50 > 1 μM], in comparison with parental cells [IC50 = 0.006 μM]), none had the T790M mutation, but showed attributes of resistance, such as up-regulation of Her-2, AXL, ERK, Akt or NF-κB in western blot analysis [20], indicating heterogeneous mechanisms of TKI resistance. PKCδ was identified as a common mediator involved in the TKI-insensitive EGFR pathways. TKI-resistant H1650 cells treated with a PKC inhibitor (sotrastaurin) or phospholipase C (PLC) inhibitor (U73122) displayed strong synergism (CI < 0.3) with gefitinib. The results demonstrated that PKCδ and PLCγ2 are implicated in the TKI-insensitive EGFR pathways [20]. PKCδ nuclear localisation was readily detectable in a TKI-resistant EGFR-mutant H1650 model. Immunofluorescence staining showed that EGFR depletion, but not kinase inhibition, reduced nuclear PKCδ. As a membrane-bound receptor, EGFR interacts with other RTKs, Her-2 and AXL, which have been previously implicated in PKCδ activation [22]. Further analysis showed that interaction between EGFR and AXL or Her-2 may sustain EGFR Y1173 phosphorylation in resistant cells treated with TKI. When phosphorylated, EGFR Y1173 functions as a docking site for PLCγ (Fig. 1a). Such findings illustrate that EGFR-Y1173-PLCγ2-nuclear PKCδ is a common axis of resistance mediated by TKI-insensitive EGFR pathways [20]. Schematic representation of the autophosphorylation sites in the EGFR and activation of the corresponding major signal transduction pathways were reported by Sordella et al. [23]. PKCδ is phosphorylated on tyrosine 374 (Y374) as a substrate of the non-receptor tyrosine phosphatase and tumour suppressor PTPN14 (Pez). Also, RIN1 (Ras and Rab interactor 1) is a binding partner of PTPN14. Loss of catalytically functional PTPN14 increases the abundance of EGFR at the cell surface of breast cancer cells. Patient survival was worse when breast cancer tissue had increased expression of the genes encoding RIN1 or PRKCD [24]. The non-RTK, feline sarcoma-related (FER), phosphorylates PKCδ on Y374 and Y374-PKCδ alters specific Ras-associated binding protein (RAB) lysosomal trafficking, which lessens RTK degradation and promotes RTK recycling [25]. Breast cancer cells with increased pY374-PKCδ levels (regulated by the opposing actions of FER and PTPN14) due to loss of PTPN14 accelerated the magnitude of ERK activation following ligand stimulation. Moreover, pY374-PKCδ inhibits Ras-associated binding protein 5 (RAB5) from late endosomes and compromises cargo degradation carried by the endosomes [25]. It was also confirmed that elevated pY374-PKCδ levels (induced by PTPN14 deficiency) led to increased cell surface expression of IGF1R and MET, as seen in EGFR tumour cells [25] (Fig. 1b). To expand the role of PKCδ in EGFR TKI resistance [20], it is tempting to hypothesise the relevance of Y374-PKCδ phosphorylation status in EGFR-mutant NSCLC cells regarding FER and PTPN14, as noted in breast cancer cells [25]. The Goodall group’s discoveries [24, 25] on dysregulation of the FER-PKCδ-PTPN14 axis and high levels of RAB5-RAB7-positive transitional endosomes in triple-negative and HER2-positive breast cancers show that phosphorylation of Y374-PKCδ stabilises the transient RAB5-RAB7-positive endosome population to shift the balance from RTK degradation to recycling. FER over-expression correlated with poor post-operative survival in NSCLC [26]. FER levels are elevated in ovarian cancer cells and loss of FER impaired the development of metastasis in ovarian cancer cells in vivo. FER phosphorylates MET at Y1349 with the activation of Src-homology 2 domain-containing phosphatase 2-mitogen-activated protein kinase (SHP2-MAPK) and RAC1-p21-activated kinase (PAK1) signalling downstream of MET in ovarian cancer cells [27]. Patients with high PAK1 mRNA or low E-cadherin mRNA respond less to TKI than patients with low PAK1 or E-cadherin tumours. It was shown that miR-145 transcription is de-repressed by phosphoinositide 3-kinase (PI3K)/Akt-mediated C/EBP-β phosphorylation, increasing PAK1 expression in gefitinib-resistant LUAD cells [28]. We carried out similar experiments in HCC827 (EGFR 19 deletion) targeting PKCι-PAK1 signalling pathways. The combination of auranofin plus OTSSP167 (MELK inhibitor) shows high synergism for inhibiting cell viability and colony formation. Mechanistically, the combination abrogates the expression of EGFR, MET, PAK1, PKCι, ERK1/2, Akt, YAP1 and mTOR. The combination was also synergistic in KRAS-mutant adenocarcinoma and squamous cell carcinoma cell lines, and decreased tumour volume in mice models [29].
a PKCδ is a common mediator involved in EGFR TKI resistance. EGFR TKIs, like gefitinib, does not inhibit EGFR pY1173 and induce the formation of inactive EGFR heterodimers. The sustained pY1173 by EGFR heterodimer promotes the activation of PLCγ2 and PKCδ and downstream oncogenic signalling pathways. EGFR interacts with other RTKs, which are implicated in PKCδ and downstream oncogenic signalling pathway activation. b Increased pY374-PKCδ levels (regulated by the opposing actions of FER and PTPN14) increase the amount of RTKs on the cell surface. EGFR epidermal growth factor receptor, TKI tyrosine kinase inhibitor, Y tyrosine, p phosphorylation, PLCγ2 phospholipase γ2, PKCδ protein kinase Cδ, SHP2 Src-homology 2 domain-containing phosphatase 2, FER feline sarcoma-related, RAB5 Ras-associated binding protein 5, RTK receptor tyrosine kinase, CDCP1 CUB domain-containing protein 1.
YAP/forkhead box protein M1 (FOXM1)
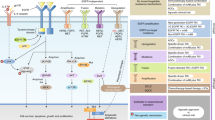
The Hippo pathway (Fig. 2) integrates multiple signals to regulate the activity of YAP and TAZ transcriptional coregulators. The primary binding partners of YAP/TAZ are transcriptionally enhanced associated domain (TEAD) transcription factors. When the Hippo pathway is off, YAP/TAZ are dephosphorylated and translocated into the nucleus, where they bind to TEAD to induce transcriptional programmes for cell proliferation and migration. Without nuclear YAP/TAZ, TEAD serves as a default repressor that binds to transcription cofactor vestigial-like protein 4, a Vg domain-containing protein [30]. Multiple tumour suppressors preserve the integrity of the Hippo pathway with cytoplasmic YAP retention, among them PTPN14 [30]. Mutations in neurofibromin 2 (NF2) have been noted in EGFR TKI-resistant NSCLC. NF2 triggers phosphorylation of YAP at S127, thereby causing cytoplasmic retention of YAP, thus preventing YAP from activating transcription of target genes. NF2 deletions have been identified by NGS in 3% of osimertinib-treated patients [17]. Upon tyrosine phosphorylation, YAP can also be activated by the Src family kinase (SFK), Yes [31] (see below).
(Left) When Hippo signalling is off, YAP enters the nucleus, interacts with TEAD and recruits other factors to induce gene transcription. (Right) When Hippo signalling is on, YAP is phosphorylated by LATS1/2 on multiple sites, resulting in interaction with 14-3-3 and cytoplasmic retention; phosphorylation also leads to YAP poly-ubiquitination and degradation. TEAD does not bind with YAP and target gene transcription is suppressed. MST1/2 mammalian Ste20-like kinase 1/2, SAV Salvador non-catalytic scaffold protein, LATS1/2 large tumour suppressor 1/2, YAP Yes-associated protein, Mob1 Mps one binder kinase activator-like 1, TEAD TEA domain family member 1, ABL Abelson murine leukaemia viral oncogene, β-Trcp β-transducin repeat-containing protein, NF2 neurofibromin 2.
Activation of the YAP and FOXM1 axis as a driver of epithelial-to-mesenchymal transition (EMT)-associated EGFR TKI resistance was discovered through an integrated transcriptomic, proteomic, and drug screening approach. Nilsson et al. (Heymach’s group) [32] found that FOXM1 is a direct transcriptional target induced by YAP. The FOXM1 protein decreased in EGFR TKI-resistant cells treated with small interfering RNA targeting YAP. Pharmacological inhibition of YAP with CA3 or verteporfin diminished FOXM1 and spindle assembly checkpoint (SAC) components, such as Aurora A, Aurora B, polo-like kinase (PLK1), and kinesin spindle protein (KSP) in erlotinib resistant cells. Along with increased nuclear localisation of YAP, EGFR TKI-resistant cells have greater sensitivity to YAP inhibition with CA3 than with the parental cells. YAP induced EGFR TKI resistance to both gefitinib and osimertinib through up-regulation of AXL [6]. In addition, AXL is often over-expressed in EGFR TKI-resistant cells and associated with mesenchymal traits, such as loss of E-cadherin and vimentin expression in several PC9 gefitinib-resistant lines [15]. Interestingly, Nilsson et al. [32] noticed that EGFR TKI-resistant cells have increased abundance of AXL, but were not sensitive to the inhibition of AXL alone, or in combination with EGFR TKIs. The Heymach group’s discoveries [32] indicate that the YAP/FOXM1 transcriptional programme up-regulates SAC components. EGFR TKI-resistant cells are highly sensitive to the inhibition of both the transcriptional pathway and SAC components (KSP, PLK1 and survivin) (Fig. 3). In addition, EGFR TKI-resistant cells were sensitive to KSP inhibitors, such as ispinesib, with IC50 values in the low nanomolar range. On the same lines, aneuploid cancer cells show increased sensitivity to genetic perturbations of core components of SAC, which ensures the proper segregation of chromosomes during mitosis. It was recently reported that aneuploid cancer cells become increasingly sensitive to inhibition of SAC over time. Aneuploid cancer cells were vulnerable to depletion of a specific mitotic kinesin, KIF8A [33, 34]. It is plausible to assume that EGFR TKI-resistant cells are aneuploid. S-phase kinase-associated protein 2 (SKP2) serves as an E3 ubiquitin-ligase repressing several proteins, including p27 and p21. YAP promotes polyploidy via Akt signalling, inducing Skp2 cytosolic retention [35]. Skp2 expression is highly up-regulated in cancers. Imipramine blue (an anti-depressant derivative) was shown to inhibit breast cancer growth by interacting and inhibiting FOXM1 and, subsequently, its transcriptional targets, including Skp2, PLK, and Aurora A, among others [36]. Furthermore, EGFR TKI-resistant cells are sensitive to Aurora A and Aurora B inhibitors [32, 37, 38]. Nanoparticle-based formulation of the Aurora B inhibitor, AZD2811, has the potential to increase efficacy and tolerability. Nilsson et al. [32] used The Cancer Genome Atlas data to assess the correlation between FOXM1 expression and SAC component gene expression in LUADs. In patients with EGFR-mutant NSCLC, FOXM1 expression was correlated with expression of PLK1 expression, AURKA/B (encoding Aurora kinase A and B), KIF11 (kinesin family member 11) (encoding KSP) and BIRC5 (baculoviral IAP repeat-containing 5) (encoding survivin) (Fig. 3). High FOXM1 expression was associated with worse disease-free survival in EGFR-mutant NSCLC. In addition, FOXM1 also enhances the activation of HGF/MET signalling extracellular signal-regulated kinase 1/2, phosphoinositide 3-kinase/Akt and signal transducer and activation of transcription 3 (STAT3) in pancreatic ductal adenocarcinoma [39] (Fig. 3). N6-methyladenosine (m6A) mRNA modification regulates mRNA splicing, export, stability and translation. ALKBH5 (α-ketoglutamarate-dependent dioxygenase homologue 5) is an RNA m6 demethylase elevated in glioblastoma stem-like cells that regulates FOXM1 (Fig. 3). The long-chain non-coding RNA (lncRNA) antisense to FOXM1 promotes the interaction of ALKBH5 with FOXM1 nascent RNA, which favours demethylation and elevated expression of FOXM1. It was shown that FOXM1 is a central mediator in glioblastoma stem-like cell proliferation [40]. Moreover, FOXM1 can act downstream of STAT3 in a ferritin-STAT3-FOXM1 feedback loop. It is noted that FOXM1 expression is observed across all subtypes of glioblastoma multiforme, while phosphorylated STAT3 is more restricted to mesenchymal subtype and absent in pro-neural glioblastoma multiforme [40].
EGFR mutations, via tyrosine phosphorylation, lead to the activation of the MAPK, AKT, STAT3 and other downstream oncogenic signalling pathways (i.e. NF-κB). SHP2 modulates signals of receptor tyrosine kinases at the level of Ras. IL-6 signals via receptor complexes, which contain gp130, and promotes STAT3 activation and nuclear translocation. gp130 associates with Src and triggers activation of YAP through phosphorylation on the tyrosine residue 357, independently of STAT3. FOXM1 is a direct transcriptional target induced by YAP. YAP/FOXM1 transcriptional programme up-regulates SAC components, AURKA/B (encoding Aurora kinase A and B) and Skp2. FOXM1, which is regulated by ALKBH5 and m6A, enhances the activation of HGF/MET (see also Fig. 1 and legend). EGFR epidermal growth factor receptor, TKI tyrosine kinase inhibitor, Y tyrosine, p phosphorylation, PLCγ2 phospholipase γ2, PKCδ protein kinase Cδ, SHP2 Src-homology 2 domain-containing phosphatase 2, FER feline sarcoma-related, RAB5 Ras-associated binding protein 5, RTK receptor tyrosine kinase, CDCP1 CUB domain-containing protein 1, YAP Yes-associated protein, IL-6 interleukin-6, ALKBH5 α-ketoglutamarate-dependent dioxygenase homologue 5, m6A N6-methyladenosine, CTGF connective tissue growth factor, SAC spindle assembly checkpoint, PLK1 polo-like kinase 1, KIF11 kinesin family member 11, BIRC5 baculoviral IAP repeat-containing 5.
Targeting YAP-mediated tumour-lineage plasticity
Plasticity between different signalling pathways, adaptive activation of bypass signalling, is inherent in temporal tumour heterogeneity under therapeutic selective pressure [41]. See the differential STAT3 phosphorylation at S727 and Y705 regulating EMT and MET processes [42] (see below). YAP/TEAD engages the EMT transcription factor SLUG to directly repress pro-apoptotic BMF. In PC9 (EGFR-mutant cells) standalone osimertinib leads to re-colonisation of wells within 8 weeks. In the combination of osimertinib with trametinib (MEK inhibitor), a few viable cells are still detected after 15 weeks of treatment. However, within days following drug withdrawal, cells proliferate and re-colonise the wells. Higher YAP/TEAD activity was observed, as measured by connective tissue growth factor (CTGF) in osimertinib-induced dormant (non-proliferative) PC9 cells. EGFR-mutant NSCLC cell lines treated for 3 weeks with osimertinib plus a tankyrase inhibitor (XAV939) reduced the number of dormant cells, diminishing regrowth. The study adds further evidence that YAP activation is necessary for cancer viability upon combined EGFR/MEK inhibition. YAP inhibition by XAV939 increases BMF (a pro-apoptotic BH3-only protein) expression in response to osimertinib in EGFR-mutant NSCLC cell lines in vitro and in vivo [43]. It was previously reported that tankyrase and the canonical Wnt pathway protect EGFR-mutant lung cancer cells from EGFR inhibition [44].
Activation of the Hippo pathway is triggered by a core kinase cascade that contains the mammalian sterile 20-like kinases (MST1/2), the large tumour suppressor 1/2 (LATS1/2) and the scaffolding protein Salvador (Sav1) (Fig. 2). Sequential phosphorylation-dependent cytoplasmic retention of the transcriptional coactivator YAP and its paralogue TAZ (transcriptional coactivator with PDZ-binding motif) occurs through interaction with 14-3-3, while the retained YAP/TAZ is downregulated by β-TrCP-mediated proteolytic degradation (Fig. 2). The regulation of YAP/TAZ subcellular localisation occurs through a variety of upstream regulators, such as Merlin, Kibra and Angiomotin, that physically bind to and exclude YAP/TAZ from the nucleus. Merlin binds to Angiomotin, releasing Rich1 (a small GTPase-activating protein) to inactivate Rac1-GTP to RAC1-GDP. In response to growth stimuli, Merlin dissociates from Angiomotin. Unoccupied Angiomotin binds to and blocks Rich1’s GAP activity, leading to increased levels of Rac1-GTP [45]. Also, PTPN14 induces cell-density-dependent nuclear export of YAP, thereby suppressing YAP oncogenic function. Mastermind-like (MAML) is a coactivator of Notch-dependent transcription that also promotes Wnt, Shh and NF-κB signalling target gene transcription. It has recently been shown that MAML1/2 also induces nuclear localisation of YAP/TAZ and enhances YAP/TAZ-TEAD-mediated transcriptional activity via the formation of a functional complex. Tissue microarray shows a strong correlation between MAML2 and YAP levels in lung cancer patients. The Cancer Genome Atlas (TCGA) data reveal a strong correlation between the mRNA levels of MAML2 and YAP target genes in lung cancer [46]. In addition, actionable YAP1-MML2 fusions have been identified in glioblastoma (AM-38), ovarian cancer (ES-2) and head and neck carcinoma (SAS) cell lines. The fusion brings together exons 1–5 of YAP1 and exons 2–5 of MAML2. YAP1-MML2 fusions are associated with increased YAP1 signalling [47]. Mir-30c contains a highly conserved region targeting MAML1 mRNA, establishing the miR-30c-MAML1-YAP/TAZ axis, which should be considered for further research [46]. Several microRNAs (miRNA), lncRNAs and circular RNAs interact with YAP and numerous key components of the Hippo pathway [48]. Zanconato et al. [49] determined that YAP/TAZ physically engages the general coactivator bromodomain-containing protein 4 (BRD4). BET inhibitors impaired the expression of YAP direct target genes, such as AXL, AURKA, FST1 and PDL1 [49]. The relevance of the YAP/TAZ-BRD4 connection in EGFR-mutant NSCLC warrants analysis.
The regulation of cytoplasmic sequestration of YAP/TAZ has also been associated with Hippo-independent mechanisms where YAP is activated by the SFK, Yes, upon tyrosine phosphorylation. The seminal work of Karin’s group [31] in inflammatory bowel diseases and colorectal cancer showed that gp130, a co-receptor for interleukin-6 (IL-6) cytokines, triggers activation of YAP and Notch, independently of the gp130 effector STAT3 [31]. The data indicate that activation of gp130 contributes to healing, regeneration and termination of inflammation via SFK-YAP-Notch signalling, in addition to the effect of STAT3 [31]. Previously, we found that gefitinib or osimertinib activates STAT3 and Src-YAP1 in EGFR-mutant lung cancer cells [5]. Over time, EGFR TKI (gefitinib) therapy led to increased mRNA expression of the YAP1 target CTGF in the treated EGFR-mutant cell population (PC9). Interestingly, combined therapy with TPCA-1 (STAT3 inhibitor) and gefitinib did not ablate this increase in CTGF expression. Gefitinib alone was unable to suppress the phosphorylation of STAT3, paxillin or YAP1 in PC9 cells. Gefitinib plus AZD0530 (SFK inhibitor) blocked paxillin and YAP1 phosphorylation on Y118 and Y357, respectively, but had no effect on pSTAT3 Y705 in PC9 cells. The triple combination of gefitinib, TPCA-1 and AZD0530 inhibited STAT3, paxillin and YAP phosphorylation [5]. Phosphorylation of YAP1 on serine 127 was not affected by any treatment, indicating that the Hippo pathway was not involved in the EGFR-mutant cell lines examined. Similar results were observed with the triple combination in H1975 cells. We hypothesised that activation of Src, acting upstream of YAP1, was a previously unrecognised event in the initial biochemical adaptation to EGFR TKI treatment in EGFR-mutant NSCLC cells (Fig. 3). Consistent with the above-mentioned studies, our previous work states that STAT3 and Src-YAP1 pathway co-activation hindered the efficacy of standalone gefitinib or osimertinib in EGFR-mutant NSCLC cells, which could be reversed by STAT3 co-inhibition [5]. We examined the mRNA levels of STAT3 and YAP1 in baseline tumour samples of 64 EGFR-mutant NSCLC patients treated with first-line EGFR TKI. Median PFS was 9.6 and 18.4 months for patients with high and low STAT3 mRNA, respectively (P < 0.001; HR for disease progression = 3.02, P = 0.001). Median PFS was 9.6 and 23.4 months for patients with high and low YAP1 mRNA, respectively (P = 0.005, HR for disease progression = 2.57, P = 0.007). Differences were also observed in median OS according to STAT3 and YAP1 mRNA expression [5]. Furthermore, according to previous reports [50], we noted that EGFR-mutant NSCLC cells co-express more than just RTKs. Non-RTKs, above all, Src, Yes and FAK, are also expressed and activated [6]. Akt and the MAPK pathways remain active, even in the presence of osimertinib [51]. EGFR, SFK and FAK concomitant inhibition enhanced the effect of osimertinib and suppressed resistance [51]. However, co-targeting EGFR and MEK [52], or Akt [15], causes growth inhibition in EGFR TKI-resistant models. The combination of osimertinib plus dasatinib was superior to the combination of osimertinib and MEK or PI3K inhibitors [51]. We demonstrated that the combination of gefitinib or osimertinib with the Src/FAK/Janus kinase 2 (JAK2) inhibitor, TPX-0005 (repotrectinib), abrogates STAT3, YAP1 and SFK activation and downregulates AXL and CDCP1 expression (Fig. 3) [6]. STAT3 was activated and translocated into the nucleus in gefitinib-treated PC9 cells, but this did not occur with TPX-0005 and the combination. We used small interfering RNA (siRNA) knockdown to address the relationship of Src-YAP1 and RTKs in EGFR-mutant NSCLC. Src, Yes or LYN siRNA reduced YAP1 phosphorylation. AXL, CDCP1 and MET phosphorylation were decreased after YAP1 or SFK knockdown, emphasising the role of YAP1 and SFKs as regulatory nodes for RTK activation [6]. We also explored the combination of EGFR TKIs with the Src inhibitor, dasatinib. Dasatinib was highly synergistic with all EGFR TKIs in PC9 and H1975 cells. However, in contrast with the Src/FAK/JAK2 inhibitor, TPX-0005, dasatinib, induced STAT3 and FAK phosphorylation. Figure 3 epitomises our conceptualisation of the interplay between RTKs and SFKs-YAP1 [6]. We have initiated a phase I study of osimertinib plus repotrectinib (formerly, TPX-0005) in EGFR-mutant NSCLC patients to test safety and response rate efficacy (TOTEM, ClinicalTrials.gov Identifier; NCT04772235).
Hippo pathway and resistance to chemotherapy
TKI-resistant EGFR-mutant cells (i.e. H1975 resistant) undergo a histological change, developing a spindle-like morphology associated with a shift of E-cadherin expression loss and appearance of vimentin, indicating EMT phenotype. An early description of individual clinical evolution in an EGFR-mutant NSCLC patient is represented by a patient with EGFR L858 LUAD and a TP53 mutation with a discreet response to first-line chemotherapy. The patient achieved near-complete response to her lung metastasis with erlotinib. After 8 months of erlotinib, a progression of lung metastases occurred. A lung core biopsy revealed the presence of EGFR T790M mutation. There was no response to chemotherapy and the patient developed bone and liver metastasis. However, a second lung core biopsy revealed the L858R EGFR mutation, but no detectable T790M mutation [53]. Compared with gefitinib alone, gefitinib combined with carboplatin plus pemetrexed improved PFS and OS, but with increased toxicity [54, 55]. Lysosomal sequestration of cytotoxic drugs and targeted therapies has been described [56]. Low lysosomal pH causes drug sequestration, impeding the release of cytotoxic agents and/or targeted agents to the cytoplasm. The effect has been reverted by verteporfin through a YAP-independent mechanism [56]. It has been reported that the Hippo-YAP pathway is involved in gemcitabine activity. Nuclear YAP enhances gemcitabine effectiveness by down-regulating multiple drug transporters and cytidine deaminase (a key enzyme that metabolises gemcitabine following its uptake). Gemcitabine treatment in cancer cell lines with Hippo pathway genetic alterations significantly reduced 3D spheroid growth, including lung cancer cells with STK11 mutations (A549, NCIH23) [57]. Conversely, intact LKB1 (STK11) enhances chemoresistance to gemcitabine by up-regulating cytidine deaminase in a basal triple-negative cell line. Also, in lung cancers with STK11 mutations, CTGF (a readout of YAP activity) has been expressed [57]. Gemcitabine in combination with platinum agents was widely used in the past, until gemcitabine was replaced by pemetrexed as the preferential chemotherapy regimen in LUAD. However, due to the role of YAP activation by Hippo-dependent or Hippo-independent mechanisms in different types of lung cancer, it is tempting to posit that gemcitabine could be a good reagent for co-treatment in EGFR-mutant NSCLC tumours with YAP over-expression, pretreatment or post-treatment. Chitinase 3 like 1 (CHI3L1) is up-regulated in most organs from COVID-19 autopsies. Chi3L1 is a secreted chitinase-like protein-modulating fibroblast proliferation, immune cell differentiation, extracellular microenvironment reorganisation and angiogenesis in response to cytokines and hypoxia [58]. High expression of CHI3L1 is associated with gemcitabine resistance in pancreatic ductal adenocarcinoma [59]. Pentoxifylline (approved for peripheral arterial disease) is an inhibitor of CHI3L1, which can revert gemcitabine resistance in pancreatic tumours [59, 60].
There is only one previous clinical study in which NSCLC EGFR-mutant patients received a chemotherapy regimen containing gemcitabine. Intercalated combination of chemotherapy and erlotinib (FASTACT-2) was a randomised study to receive six cycles of gemcitabine (1250 mg/m2 on days 1 and 8, intravenously), plus platinum (carboplatin 5 × area under the curve) or cisplatin (75 mg/m2 on day 1, intravenously) with intercalated erlotinib (chemotherapy plus erlotinib, 150 mg/day on days 15–28, orally), or placebo (chemotherapy plus placebo, orally), every 4 weeks. Treatment benefit was noted only in patients with EGFR mutations. Median PFS was 16.8 versus 6.9 months, and median OS was 31.4 versus 20.6 months [61]. Although the standard of care recommends pemetrexed and carboplatin in LUAD patients, the use of gemcitabine requires further investigation in EGFR-mutant NSCLC. Bear in mind that chemotherapy, per se, promotes immune evasion phenotype in surviving triple-negative breast cancer cells. Exposure of triple-negative breast cancer cells to chemotherapy (carboplatin, doxorubicin, gemcitabine or paclitaxel) induces the expression of HIF-1α and HIF-2α, leading to the expression of PD-L1, CD73 and CD47, which promotes suppression of innate antitumour immunity mediated by macrophages, dendritic cells and myeloid-derived suppressor cells, as well as suppression of adaptive immunity mediated by T cells [62]. CDT3 expression is also increased in EGFR-mutant NSCLC. Using a fold change >1.2, 15 genes were up-regulated in EGFR-mutant tumours in the TCGA cohort. The top up-regulated gene was ADORA1 (up-regulation fold change = 2.16, P < 0.001) encoding the adenosine receptor 1, and the sixth up-regulated gene was NT5E (up-regulation fold change = 1.44, P = 0.02) encoding CD73, both of which are key molecules in the CD73/adenosine pathway. An ongoing trial combining oleclumab (anti-CD73) with osimertinib or oleclumab and AZD4635 (adenosine receptor inhibitor) (NCT03381274) evaluates the CD73/adenosine pathway blockade in EGFR-mutant NSCLC [63]. It was seen that inhibition of ADORA1 induces the up-regulation of PD-L1 via transcription factor ATF3 in melanoma and NSCLC. ADORA1 and ATF3 levels predicted the efficacy of PD-1 immune checkpoint inhibitors [64]. Ataxia-telangiectasia mutated (ATM)-associated DNA-damage-induced T cell senescence and dysfunction by both regulatory T cells and tumour cells. The study indicates that ATM-associated DNA-damage initiation and MAPK and STAT3 signalling activation control lipid metabolism in senescent T cells in vitro. Blockade of ATM activation with the ATM-specific inhibitor, KU55933, decreased mRNA and protein expression of group IV phospholipase A2α in regulatory T-induced senescent T cells [65].
Neurotransmitters in EGFR-mutant NSCLC
Monique Nilsson and John Heymach [66] reported that stress hormones (norepinephrine) activate β2-adrenergic receptors on NSCLC cells that cooperate with mutant EGFR, resulting in the inactivation of LKB1 and subsequent secretion of IL-6. β2-Adrenergic (ADRB2) activation, with tumour growth and EGFR TKI resistance, was abrogated with β-blockers or IL-6 inhibitors. Propranolol (β-blocker) blocked norepinephrine-induced IL-6. We found synergism with carvedilol (another β-blocker in clinical practice), but not with propranolol in EGFR-mutant resistant cells pretreated with norepinephrine (unpublished). mRNA expression of ADRB2 was elevated in NSCLC patients, as well as in NSCLC cell lines. Elevated IL-6 determined by enzyme-linked immunosorbent assay (ELISA) was associated with worse prognosis in EGFR TKI-treated NSCLC patients, while β-blocker use was associated with lower IL-6 concentrations and better benefit from EGFR TKIs [66]. Recently, it has been reported that a gut microbe formed phenylacetyl glutamine that activates β-adrenergic receptors on platelets in experimental mice models [67]. Presumably, multi-factorial stimuli can converge in up-regulating β-adrenergic receptors. Norepinephrine-ADRB2 signalling up-regulates nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) in pancreatic cancer models [68]. ADRB2 blockade, together with gemcitabine, reduced NGF expression and increased survival in pancreatic cancer (βKRAS+/G12D; LSL-Trp53+/R172H; Pdx-Cre [KPC]) mice [68]. BDNF expression was high in pancreatic cancer patients and survival was increased in patients receiving non-selective β-blockers. TrkA (neurotrophic tyrosine kinase receptor 1 (NTRK1)) is the receptor for NGF and TrkB (NTRK2) is the receptor for BDNF. It was found that blocking ADRB2 and/or Trk receptors was effective in the experimental pancreatic models [68]. In NSCLC, BDNF also activates TrkB and STAT3 signalling, both in cancer cell lines and in lung cancer tissues, where the levels of pSTAT3 Y705 and pTrkB were detected by immunoblotting [69]. Blocking TrKB with the Trk inhibitor, K252a, decreased STAT3 phosphorylation at Y705. TrkB expression has been detected in plasma exosomes of glioblastoma patients. The study showed that YKL-40 (CHI3L1) and YKL-40-silenced cells induced a decrease in TrkB [70]. Such findings can impinge upon further research in EGFR-mutant NSCLC cells. In addition, TrkA and NGF have been shown to be more increased in lung squamous cell carcinoma than in adenocarcinoma [71]. Squamous cell transformation has been described as a mechanism of resistance to osimertinib [72]. An early study identified that the NTKRs, TrkA and TrkB, are involved in resistance to TKI in EGFR-mutant PC9 cells and that pharmacological blockade of TrkA with lestaurtinib restored sensitivity to erlotinib. Other EGFR bypassing kinases were also identified [73]. TrkA (a.k.a. NGF) RTK (NTRK1) induces YAP activation by suppressing LATS in pancreatic and breast cancer cell lines [74]. CTGF (a.k.a CCN2) (a readout of YAP1 activation) interacts with TrKA and EGFR/TrkA receptor. Crosstalk was found in response to CTGF stimulation [75]. It has been seen that glioma cells produce large amounts of CTGF, which activates the CTGF-EGFR pathway of glioma cells [76].
STAT3, IL-6 and leukaemia inhibitory factor (LIF)
STAT3 is a latent transcription factor found in the cytoplasm and is activated by tyrosine phosphorylation, leading to dimerisation and nuclear translocation [77]. We adopted immunofluorescence to track STAT3 activation state by its intracellular localisation. STAT3 was activated and translocated into the nucleus with gefitinib, but this did not occur with TPX-0005 and the combination [6]. STAT3 Y705 phosphorylation occurs a few hours after treatment with EGFR TKIs inducing surviving tumour cells to adapt to a quiescent, dormant persistent state [78]. We noted that afatinib equally causes phosphorylation of STAT3 Y705 and elevation of STAT3 and RANTES (regulated upon activation, normal T cell expressed and presumably secreted) mRNA levels. Aldehyde dehydrogenase-positive cells were observed and HES1 (Hairy and Enhancer of Split 1) was elevated following therapy with afatinib plus TPCA-1 (STAT3 inhibitor) [79]. As previously observed [77], we depicted a model showing that EGFR-mutant lung cancer cells produce high IL-6 levels, which subsequently activates the gp130/JAK/STAT3 pathway. Unphosphorylated STAT3 (U-STAT3) activates genes, such as RANTES, when induced by the activation of STAT3 in response to EGFR mutations or ligands (i.e. IL-6). Although afatinib inhibits ERK and Akt signalling in PC9, PC9-GR2 (AXL over-expression) and PC9-GR4 (38% allelic fraction of T790M), it does not abolish phosphorylation of STAT3 T705. Also, a gradual increase in STAT3 and RANTES mRNA levels was observed after 7 days of treatment. Jacqueline Bromberg’s group [77] examined mRNA levels of IL-6, OSM (oncostatin M), LIF, IL-11 and CNTF (ciliary neurotrophic factor), but only IL-6-mRNA was detected in 11–18, H3255, H1650 and H1975 cell lines. IL-6-secreted protein levels were determined by ELISA in the conditioned medium collected for near-confluent cell cultures: 11–18 cells, 1800 pg/ml; H3255 cells, 2900 pg/ml; H1650 cells, 7700 pg/ml; and H1975 cells, 7800 pg/ml. These levels were higher in comparison with levels of normal cells or other cancer-derived cell lines for which no >10–60 pg/ml were seen [77]. These data, together with other experiments, helped to discover that IL-6 is secreted by EGFR-mutant NSCLC cells, leading to the activation of the gp130/JAK/STAT3 signalling pathway [77].
LIF receptor (LIFR) is a well-recognised upstream regulator of the Hippo pathway. LIFR expression suppresses nuclear YAP disposition in breast cancer cells. LIFR expression status correlates with PFS in breast cancer patients [80]. Intriguingly, LIF activates LIFR/p-ERK/STAT3 S727 phosphorylation in MET process in lung cancer. The findings provide a new glimpse of the putative processes in EMT/MET in EGFR-mutant NSCLC. By studying metastasis in bone marrow-derived mesenchymal stem cell (BM-MSC)-driven lung cancer models, it was shown that BM-MSCs elicit EMT in epithelial-type cells through the IL-6/STAT3 Y705 phosphorylation pathway while inducing MET in mesenchymal-type cells through LIFR/STAT3 S727 phosphorylation [42]. These findings complement the Bromberg group’s seminal discoveries [77]. STAT3 phosphorylation at Y705 and S727 is a readout of EMT and MET fluctuations according to the tumour evolution and adaptive mechanisms of treatment resistance. Such a concept warrants further analysis in EGFR-mutant NSCLC and could provide a new means for gauging the plasticity of EGFR-mutant cancer cells. It is tempting to speculate that the levels of IL-6 and LIF could be surrogate markers in the evolution of EGFR-mutant NSCLC.
Further convolution in EGFR TKI resistance (tolerance) is the role of SOX2 and TGFβ signalling [81,82,83,84]. As aforementioned, TKI therapy favoured mesenchymal traits in lung cancer cells, with deficient SOX2 expression, whereas SOX2 expression promotes TKI sensitivity and inhibited the mesenchymal phenotype [81]. SOX2 belongs to the SOX (Syr-related HMG Box) family of proteins and responds to respiratory tract injuries. SOX2 signalling initiates the proliferation and differentiation of lung progenitor cells to maintain tissue homeostasis. SOX2, in conjunction with OCT4, KLF4 and MYC, can reverse the mesenchymal morphology of fibroblasts and reprogramme them into pluripotent stem cells. Inhibition of TGβ signalling facilitates the SOX2-mediated reprogramming process of fibroblasts [81]. Tumours expressing low SOX2 and high vimentin signature were associated with worse survival outcomes in EGFR-mutant patients [81]. Moreover, TGFBR1/2 receptors were up-regulated in HCC827 GR and H1975 AZDR-resistant lines. Epigenetic modifications of H3K27ac and H3K4m3 (which marks the active enhancer and promoter, respectively) were higher in the SOX2 locus in HCC827 cells compared with its TKI-tolerant counterpart, HCC827 GR. The study clearly shows that the loss of SOX2 expression by TGFβ switches off SOX2-EGFR signalling and induces EMT with decreased BCL2L11 (Bim) pro-apoptotic signalling; henceforth, increasing EGFR TKI tolerance [81].
Concurrent alterations in EGFR-mutant NSCLC
Targeted NGS performed by Helena Yu and investigators at Memorial Sloan-Kettering Cancer Center in 200 EGFR-mutant pretreatment samples revealed that the most frequent concomitant genetic defects were mutations in TP53, PIK3CA (phosphatidylinositol-4-5-bisphosphate 3-kinase catalytic subunit alpha), CTNB1 and RB1, and focal amplifications in EGFR, TTF1, MDM2, CDK4 and FOXA1. Moreover, shorter PFS with EGFR TKI was related to the amplification of ERBB2 or MET, or mutations in TP53. In post-treatment samples, in addition to EGFR T790M (51%), MET (7%), BRAF fusion, FGFR3 fusion, YES amplification and KEAP loss were also detected [85]. Other primal studies showed the bonafide of outlining co-existing genetic defects in EGFR-mutant NSCLC using circulating tumour DNA (ctDNA)-targeted NGS assays [86]. The co-occurrence of TP53 mutations is high, 65–75%, in ctDNA or tumour samples, respectively, and correlates with shorter PFSl [87,88,89,90].
Patients with EGFR/RB1/TP53-mutant NSCLC represented 5% (43 of 863) of EGFR-mutant NSCLC and, irrespective of the risk of small cell lung cancer transformation, patients with EGFR/TP53/RB1 mutations had a shorter time to discontinuation than EGFR/TP53 and EGFR-mutant only cancers (9.5 versus 12.3 versus 36.6 months, respectively) [90]. Arsenic trioxide (ATO) used for the treatment of acute promyelocytic leukaemia can rescue multiple p53 mutants. ATO inhibits the growth of cell lines with p53 mutations at structural hotspots in five lines: RFX 393 (R175H), CCRF-CEM (R175H R248Q), Hop-92 (R175L), Sk-MEL-2 (245S) and BT-549 (R249S). ATO did not significantly reactivate any DNA-contact p53 mutants. In contrast, the structural p53 mutations, V272M, R282W, E285K and Y234C, were rescued by ATO in transactivation. This study highlights the opportunity of repurposing ATO in treating p53-mutant cancer patients [91].
Patients with concurrent EGFR/TP53/RB1 alterations also displayed gene defects in PIK3CA (20%), NRTK1 (11%), MCL1 (11%), NK2 homeobox 1 (11%), ERBB2 (9%), FOXA1 (9%), PLCγ2 (9%), PTEN (9%), RNA-binding motif protein 10 (RBM10) (9%) and others [90]. Activation-induced cytidine deaminase/apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like APOBEC mutation was also enriched in EGFR/TP53/RB1-mutant NSCLC [90]. Intriguingly, APOBEC3B (A3B) cytosine deaminase is repressed by p53, while p53 mutation increases A3B expression. In addition, cytotoxic agents such as cisplatin, etoposide or 5-fluorouracil induced A3B expression and cytosine deamination. Chemotherapy induction of A3B expression is directed by DNA-PKcs/ATM/Akt activation of NF-κB. In T47D cells, the DNA-PKcs inhibitor, NU7411, inhibited A3B expression in a dose-dependent manner. In all cancer cell lines tested, chemotherapy drugs stimulated A3B expression and DNA-PK, ATM, Akt or NF-κB inhibitors reduced cisplatin-induced A3B expression [92].
Future perspectives
LUAD driven by EGFR mutations is one of the most recognised and studied types of non-small cell lung cancer, specifically at the research level in human EGFR-mutant cell lines, and at the clinical level in pharmaceutical industry-oriented clinical trials. This review stresses basic key points accumulated in laboratory research, with the goal of making the information accessible and translatable to EGFR-mutant patients, both as novel biomarkers and as combinatory therapies for improving survival in stage IV EGFR-mutant LUAD patients. NGS assays are ineludibly necessary for management guidance; however, at present, co-mutational landscape information is merely informative since no therapies are currently available for patients with commonly co-occurring TP53 mutations. In the authors’ opinion, further endeavours should be made to yield patient-derived organoids, as they could offer a wide opportunity to better understand the cancer cell dynamics of each individual patient beyond the standardisation of care.
References
Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90.
Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–18.
Sun X, Song Q, He L, Yan L, Liu J, Zhang Q, et al. Receptor tyrosine kinase phosphorylation pattern-based multidrug combination is an effective approach for personalized cancer treatment. Mol Cancer Ther. 2016;15:2508–20.
Gusenbauer S, Vlaicu P, Ullrich A. HGF induces novel EGFR functions involved in resistance formation to tyrosine kinase inhibitors. Oncogene. 2013;32:3846–56.
Chaib I, Karachaliou N, Pilotto S, Codony Servat J, Cai X, Li X, et al. Co-activation of STAT3 and YES-associated protein 1 (YAP1) pathway in EGFR-mutant NSCLC. J Natl Cancer Inst. 2017;109:1–12.
Karachaliou N, Chaib I, Cardona AF, Berenguer J, Bracht JWP, Yang J, et al. Common co-activation of AXL and CDCP1 in EGFR-mutation-positive non-small cell lung cancer associated with poor prognosis. EBioMedicine. 2018;29:112–27.
Taniguchi H, Yamada T, Wang R, Tanimura K, Adachi Y, Nishiyama A, et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat Commun. 2019;10:259.
Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–58.
Tomasello C, Baldessari C, Napolitano M, Orsi G, Grizzi G, Bertolini F, et al. Resistance to EGFR inhibitors in non-small cell lung cancer: Clinical management and future perspectives. Crit Rev Oncol Hematol. 2018;123:149–61.
Nair A, Chung HC, Sun T, Tyagi S, Dobrolecki LE, Dominguez-Vidana R, et al. Combinatorial inhibition of PTPN12-regulated receptors leads to a broadly effective therapeutic strategy in triple-negative breast cancer. Nat Med. 2018;24:505–11.
Davra V, Kumar S, Geng K, Calianese D, Mehta D, Gadiyar V, et al. Axl and Mertk receptors cooperate to promote breast cancer progression by combined oncogenic signaling and evasion of host anti-tumor immunity. Cancer Res. 2020. https://doi.org/10.1158/0008-5472.CAN-20-2066.
Du W, Zhu J, Zeng Y, Liu T, Zhang Y, Cai T, et al. KPNB1-mediated nuclear translocation of PD-L1 promotes non-small cell lung cancer cell proliferation via the Gas6/MerTK signaling pathway. Cell Death Differ. 2021;28:1284–1300.
Ma S, Zhang L, Ren Y, Dai W, Chen T, Luo L, et al. Epiregulin confers EGFR-TKI resistance via EGFR/ErbB2 heterodimer in non-small cell lung cancer. Oncogene. 2021;40:2596–609.
Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60.
Jacobsen K, Bertran-Alamillo J, Molina MA, Teixido C, Karachaliou N, Pedersen MH, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun. 2017;8:410.
Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–6.
Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res. 2020;26:2654–63.
Tumbrink HL, Heimsoeth A, Sos ML. The next tier of EGFR resistance mutations in lung cancer. Oncogene. 2021;40:1–11.
Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121:725–37.
Lee PC, Fang YF, Yamaguchi H, Wang WJ, Chen TC, Hong X, et al. Targeting PKCδ as a therapeutic strategy against heterogeneous mechanisms of EGFR inhibitor resistance in EGFR-mutant lung cancer. Cancer Cell. 2018;34:954–.e954.
Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–61.
Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, et al. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533–46.
Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7.
Belle L, Ali N, Lonic A, Li X, Paltridge JL, Roslan S, et al. The tyrosine phosphatase PTPN14 (Pez) inhibits metastasis by altering protein trafficking. Sci Signal. 2015;8:ra18.
Lonic A, Gehling F, Belle L, Li X, Schieber NL, Nguyen EV, et al. Phosphorylation of PKCdelta by FER tips the balance from EGFR degradation to recycling. J Cell Biol. 2021;220:1–22.
Kawakami M, Morita S, Sunohara M, Amano Y, Ishikawa R, Watanabe K, et al. FER overexpression is associated with poor postoperative prognosis and cancer-cell survival in non-small cell lung cancer. Int J Clin Exp Pathol. 2013;6:598–612.
Fan G, Zhang S, Gao Y, Greer PA, Tonks NK. HGF-independent regulation of MET and GAB1 by nonreceptor tyrosine kinase FER potentiates metastasis in ovarian cancer. Genes Dev. 2016;30:1542–57.
Wu DW, Wu TC, Chen CY, Lee H. PAK1 is a novel therapeutic target in tyrosine kinase inhibitor-resistant lung adenocarcinoma activated by the PI3K/AKT signaling regardless of EGFR mutation. Clin Cancer Res. 2016;22:5370–82.
Ito M, Codony-Servat C, Codony-Servat J, Llige D, Chaib I, Sun X, et al. Targeting PKCiota-PAK1 signaling pathways in EGFR and KRAS mutant adenocarcinoma and lung squamous cell carcinoma. Cell Commun Signal. 2019;17:137.
Ma S, Meng Z, Chen R, Guan KL. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604.
Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62.
Nilsson MB, Sun H, Robichaux J, Pfeifer M, McDermott U, Travers J, et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12:eaaz4589.
Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021. https://doi.org/10.1038/s41586-020-03114-6.
Quinton RJ, DiDomizio A, Vittoria MA, Kotynkova K, Ticas CJ, Patel S, et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature. 2021. https://doi.org/10.1038/s41586-020-03133-3.
Zhang S, Chen Q, Liu Q, Li Y, Sun X, Hong L, et al. Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell. 2017;31:669–84.e667.
Rajamanickam S, Panneerdoss S, Gorthi A, Timilsina S, Onyeagucha B, Kovalskyy D, et al. Inhibition of FoxM1-mediated DNA repair by Imipramine blue suppresses breast cancer growth and metastasis. Clin Cancer Res. 2016;22:3524–36.
Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–8.
Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Gimenez-Capitan A, Cantero F, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10:1812.
Cui J, Xia T, Xie D, Gao Y, Jia Z, Wei D, et al. HGF/Met and FOXM1 form a positive feedback loop and render pancreatic cancer cells resistance to Met inhibition and aggressive phenotypes. Oncogene. 2016;35:4708–18.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 EXPRESSION AND CELL PROLIFERATION PROGRam. Cancer Cell. 2017;31:591–606.e596.
Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94.
Lin WH, Chang YW, Hong MX, Hsu TC, Lee KC, Lin C, et al. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT-MET switch and cancer metastasis. Oncogene. 2021;40:791–805.
Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, et al. Treatment-induced tumor dormancy through yap-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–22.e112.
Casas-Selves M, Kim J, Zhang Z, Helfrich BA, Gao D, Porter CC, et al. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res. 2012;72:4154–64.
Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–40.
Kim J, Kwon H, Shin YK, Song G, Lee T, Kim Y, et al. MAML1/2 promote YAP/TAZ nuclear localization and tumorigenesis. Proc Natl Acad Sci USA. 2020;117:13529–40.
Picco G, Chen ED, Alonso LG, Behan FM, Goncalves E, Bignell G, et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun. 2019;10:2198.
Liu C, Wu Y, Ma J. Interaction of non-coding RNAs and Hippo signaling: Implications for tumorigenesis. Cancer Lett. 2020;493:207–16.
Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med. 2018;24:1599–610.
Yoshida T, Zhang G, Smith MA, Lopez AS, Bai Y, Li J, et al. Tyrosine phosphoproteomics identifies both codrivers and cotargeting strategies for T790M-related EGFR-TKI resistance in non-small cell lung cancer. Clin Cancer Res. 2014;20:4059–74.
Ichihara E, Westover D, Meador CB, Yan Y, Bauer JA, Lu P, et al. SFK/FAK Signaling Attenuates Osimertinib Efficacy in Both Drug-Sensitive and Drug-Resistant Models of EGFR-Mutant Lung Cancer. Cancer Res. 2017;77:2990–3000.
Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, et al. Combined EGFR/MEK inhibition prevents the emergence of resistance in EGFR-mutant. Lung Cancer Cancer Discov. 2015;5:960–71.
Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26.
Hosomi Y, Morita S, Sugawara S, Kato T, Fukuhara T, Gemma A, et al. Gefitinib alone versus gefitinib plus chemotherapy for non-small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 Study. J Clin Oncol. 2020;38:115–23.
Noronha V, Patil VM, Joshi A, Menon N, Chougule A, Mahajan A, et al. Gefitinib versus gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J Clin Oncol. 2020;38:124–36.
Gavini J, Dommann N, Jakob MO, Keogh A, Bouchez LC, Karkampouna S, et al. Verteporfin-induced lysosomal compartment dysregulation potentiates the effect of sorafenib in hepatocellular carcinoma. Cell Death Dis. 2019;10:749.
Gujral TS, Kirschner MW. Hippo pathway mediates resistance to cytotoxic drugs. Proc Natl Acad Sci USA. 2017;114:E3729.
Nie X, Qian L, Sun R, Huang B, Dong X, Xiao Q, et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184:775–91.e714.
Xavier CPR, Castro I, Caires HR, Ferreira D, Cavadas B, Pereira L, et al. Chitinase 3-like-1 and fibronectin in the cargo of extracellular vesicles shed by human macrophages influence pancreatic cancer cellular response to gemcitabine. Cancer Lett. 2021;501:210–23.
Kim JH, Shin BC, Park WS, Lee J, Kuh HJ. Antifibrotic effects of pentoxifylline improve the efficacy of gemcitabine in human pancreatic tumor xenografts. Cancer Sci. 2017;108:2470–7.
Wu YL, Lee JS, Thongprasert S, Yu CJ, Zhang L, Ladrera G, et al. Intercalated combination of chemotherapy and erlotinib for patients with advanced stage non-small-cell lung cancer (FASTACT-2): a randomised, double-blind trial. Lancet Oncol. 2013;14:777–86.
Samanta D, Park Y, Ni X, Li H, Zahnow CA, Gabrielson E, et al. Chemotherapy induces enrichment of CD47(+)/CD73(+)/PDL1(+) immune evasive triple-negative breast cancer cells. Proc Natl Acad Sci USA. 2018;115:E1239–48.
Le X, Negrao MV, Reuben A, Federico L, Diao L, McGrail D, et al. Characterization of the immune landscape of EGFR-mutant NSCLC identifies CD73/adenosine pathway as a potential therapeutic target. J Thorac Oncol. 2021;16:583–600.
Liu H, Kuang X, Zhang Y, Ye Y, Li J, Liang L, et al. ADORA1 inhibition promotes tumor immune evasion by regulating the ATF3-PD-L1 axis. Cancer Cell. 2020;37:324–39.e328.
Liu Y, Liu G, Wang J, Zheng Z-y, Jia L, Rui W, et al. Chimeric STAR receptors using TCR machinery mediate robust responses against solid tumors. Sci Transl Med. 2021;13:eabb5191.
Nilsson MB, Sun H, Diao L, Tong P, Liu D, Li L, et al. Stress hormones promote EGFR inhibitor resistance in NSCLC: implications for combinations with β-blockers. Sci Transl Med. 2017;9:1–10.
Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell. 2020;180:862–77.e822.
Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, et al. Beta2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 2018;33:75–90.e77.
Chen B, Liang Y, He Z, An Y, Zhao W, Wu J. Autocrine activity of BDNF induced by the STAT3 signaling pathway causes prolonged TrkB activation and promotes human non-small-cell lung cancer proliferation. Sci Rep. 2016;6:30404.
Pinet S, Bessette B, Vedrenne N, Lacroix A, Richard L, Jauberteau MO, et al. TrkB-containing exosomes promote the transfer of glioblastoma aggressiveness to YKL-40-inactivated glioblastoma cells. Oncotarget. 2016;7:50349–64.
Gao F, Griffin N, Faulkner S, Rowe CW, Williams L, Roselli S, et al. The neurotrophic tyrosine kinase receptor TrkA and its ligand NGF are increased in squamous cell carcinomas of the lung. Sci Rep. 2018;8:8135.
Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res. 2020. https://doi.org/10.1158/1078-0432.CCR-19-3563
Sharifnia T, Rusu V, Piccioni F, Bagul M, Imielinski M, Cherniack AD, et al. Genetic modifiers of EGFR dependence in non-small cell lung cancer. Proc Natl Acad Sci USA. 2014;111:18661.
Yang X, Shen H, Buckley B, Chen Y, Yang N, Mussell AL, et al. NTRK1 is a positive regulator of YAP oncogenic function. Oncogene. 2019;38:2778–87.
Rayego-Mateos S, Rodrigues-Diez R, Morgado-Pascual JL, Rodrigues Diez RR, Mas S, Lavoz C, et al. Connective tissue growth factor is a new ligand of epidermal growth factor receptor. J Mol Cell Biol. 2013;5:323–35.
Xu X, Liu Y, Li Y, Chen H, Zhang Y, Liu J, et al. Selective exosome exclusion of miR-375 by glioma cells promotes glioma progression by activating the CTGF-EGFR pathway. J Exp Clin Cancer Res. 2021;40:16.
Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–56.
Fan W, Tang Z, Yin L, Morrison B, Hafez-Khayyata S, Fu P, et al. MET-independent lung cancer cells evading EGFR kinase inhibitors are therapeutically susceptible to BH3 mimetic agents. Cancer Res. 2011;71:4494–505.
Codony-Servat C, Codony-Servat J, Karachaliou N, Molina MA, Chaib I, Ramirez JL, et al. Activation of signal transducer and activator of transcription 3 (STAT3) signaling in EGFR mutant non-small-cell lung cancer (NSCLC). Oncotarget. 2017;8:47305–16.
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–7.
Kuo MH, Lee AC, Hsiao SH, Lin SE, Chiu YF, Yang LH, et al. Cross-talk between SOX2 and TGFbeta signaling regulates EGFR-TKI tolerance and lung cancer dissemination. Cancer Res. 2020;80:4426–38.
Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–50.
Rosell R. Mediating resistance in oncogene-driven cancers. N Engl J Med. 2013;368:1551–2.
Wang C, Wang T, Lv D, Li L, Yue J, Chen HZ, et al. Acquired resistance to EGFR TKIs mediated by TGFbeta1/integrin beta3 signaling in EGFR-mutant lung cancer. Mol Cancer Ther. 2019;18:2357–67.
Yu HA, Suzawa K, Jordan E, Zehir A, Ni A, Kim R, et al. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24:3108–18.
Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49:1693–704.
Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6:681–90.
Duan J, Xu J, Wang Z, Bai H, Cheng Y, An T, et al. Refined stratification based on baseline concomitant mutations and longitudinal circulating tumor DNA monitoring in advanced EGFR-mutant lung adenocarcinoma under gefitinib treatment. J Thorac Oncol. 2020;15:1857–70.
Kim Y, Lee B, Shim JH, Lee SH, Park WY, Choi YL, et al. Concurrent genetic alterations predict the progression to target therapy in EGFR-mutated advanced NSCLC. J Thorac Oncol. 2019;14:193–202.
Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol. 2019;14:1784–93.
Chen S, Wu JL, Liang Y, Tang YG, Song HX, Wu LL, et al. Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell. 2021;39:225–39 e228.
Periyasamy M, Singh AK, Gemma C, Farzan R, Allsopp RC, Shaw JA, et al. Induction of APOBEC3B expression by chemotherapy drugs is mediated by DNA-PK-directed activation of NF-kappaB. Oncogene. 2021;40:1077–90.
Acknowledgements
Figures in this manuscript were created with BioRender.com.
Funding
Dr. RR reports a grant from the Spanish Association Against Cancer, AECC (PROYE18012ROSE), and funding from a European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement ELBA No. 765492.
Author information
Authors and Affiliations
Contributions
Writing, review and/or revision of the manuscript: RR, AFC, OA, AA, MI, CP, JC-S and MS.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rosell, R., Cardona, A.F., Arrieta, O. et al. Coregulation of pathways in lung cancer patients with EGFR mutation: therapeutic opportunities. Br J Cancer 125, 1602–1611 (2021). https://doi.org/10.1038/s41416-021-01519-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01519-2
This article is cited by
-
Direct and selective pharmacological disruption of the YAP–TEAD interface by IAG933 inhibits Hippo-dependent and RAS–MAPK-altered cancers
Nature Cancer (2024)
-
HER3 functions as an effective therapeutic target in triple negative breast cancer to potentiate the antitumor activity of gefitinib and paclitaxel
Cancer Cell International (2023)