Abstract
Sophoridine is a quinolizidine natural product and the exploration of its derivatives has been carried out, and the potent anticancer compound IMB-HDC was acquired. Although previous studies have revealed that some sophoridine derivatives could induce DNA breakage, the underlying mechanisms of inhibition of DNA damage repair (ATR inactivation) and the apoptosis independent of p53, have not been elucidated. Our research reveals a novel DNA response mechanism different from general DNA-damaging agents, and that sophoridine derivate inhibits the phosphorylation of Tyr694 and Ser780 of STAT5a to induce the lessened shuttle from the cytoplasm to the nucleus, and leads to the decreased nuclear STAT5a and subsequently inhibits the expression of STAT5a target gene RAD51 that contributes to the checkpoint activation, thus inhibiting ATR activation. Meanwhile, IMB-HDC that induced the diminished expression of STAT5a target gene contributes to proliferation and leads to apoptosis. More importantly, we give the first evidence that promoting the effect of Tyr694 phosphorylation on nuclear location and subsequent STAT5a target gene transcription depends on Ser780 increased or unchanged phosphorylation and was not correlated with Ser726 phosphorylation.
Similar content being viewed by others
Introduction
Sophoridine (Fig. 1a, left) was a quinolizidine natural product and was abstracted from sophora alopecuroides. It plays an important role in anti-inflammatory [1, 2], antivirus, and improvement of the cardiac action [3]. Its antitumor activity was ascertained in 1977, and in 1993 sophoridine acquired the approval of clinical study [4, 5]. In 2005, sophoridine injection was approved by Chinese FDA and used to treat malignant trophoblastic tumors [5]. Although sophoridine as the anticancer agent was approved, its anticancer effect was only observed in malignant trophoblastic tumors. To improve the anticancer effect of sophoridine, a series of structure modifications based on sophoridine were performed, and the anticancer activities of the corresponding derivatives were examined. Among these derivatives, sophoridinic acid with a three-ring structure scaffold displayed the potent antiproliferative activities compared with their parent and other derivatives. Not only could three-ring structure compounds exert potent inhibitory effect on malignant trophoblastic tumor, but could also inhibit the growth of other tumor cell lines effectively, such as hepatic carcinoma, colon cancer, pancreatic cancer, etc. [6, 7].
a Structure of sophoridinol and IMB-HDC. b The cell morphology of HCT-8 cells was observed at 0, 3, 7, and 24 h after treatment with 5 μM IMB-HDC. c K562, HCT-8, and HepG2 cells were treated with the indicated concentrations of IMB-HDC, and cell survival was detected by the SRB assay. The dose-dependent curves were depicted. d About 3 μM IMB-HDC was added into HCT-8 cells, MIA-PaCa-2 cells, and HepG2 cells for 24, 48, and 72 h, and cell vitality was examined, and time-dependent curves were plotted. e HCT-8 cells and MIA-PaCa-2 cells were seeded in a six-well plate, after 24 h, various concentrations of IMB-HDC were added and continued to be incubated with cancer cells for 7 days, and then visible colonies were counted as described in the “Materials and methods” section. Colony-formation rate = (number of colonies/number of seeded cells) × 100%. f IC50 of IMB-HDC on various tumor cells. g Various cells were treated with sophoridine or IMB-HDC for 48 h, and cell vitality was measured. Scale bar represents 25 μm. Mean±SD. *P < 0.05; **P < 0.01.
Based on sophoridinic acid with three-ring structure, we made a series of modifications and acquired the associated derivatives. Subsequent anticancer activity screening showed that the novel structure compound IMB-HDC (Fig. 1a) possessed the stronger anticancer activity compared with its parent or other structure derivatives with three-ring structure. In addition, compared with other derivatives, IMB-HDC also shows good water solubility, lower toxicity, and synthesizes easily, suggesting that it is worthy to be further explored as the anticancer compound.
Although many studies on the anticancer mechanisms of sophoridine or its derivatives have been performed, they could induce DNA breakage and cell apoptosis and these changes were independent of p53, and found that DNA damage response was inhibited (such as ATR inactivation); although p53 and ATR activation were vital in DNA damage repair (DDR) [7], the underlying molecular mechanisms of these aberrant events different from conventional DNA-damaging compounds had not been elucidated clearly.
Stat5a is a key transcription factor and signaling protein that mediates diverse cellular responses to cytokines and growth factors [8]. In response to cytokines and growth factors, STAT5a is phosphorylated by the receptor-associated kinases, and then forms homo- or heterodimers that translocate to the cell nucleus where they act as the transcription activator [8, 9]. STAT5a could trigger the transcription of some genes such as Bcl-2, ClycinD1, Pim, c-fos, and β-catenin to regulate proliferation and apoptosis. In addition to promoting proliferation activity, STAT5a also revealed that it participated in the DDR, and it could upregulate the expression of DDR genes, such as RAD51, a vital component in homologous recombination of DNA during double-strand break repair, and its deletion could induce checkpoint inactivation to subsequently suppress ATR activation [9]. Generally, as a target molecule of p53 in response to DDR, STAT5a expression increased as a result of p53 stability and then plays the promoting repair of DNA damage in DDR.
Given that the biological effect of STAT5a was dependent on its target gene expression, STAT5a nuclear location was the key to the transcriptional initiation of its target gene and biological function preformation. Previous reports showed that tyrosine phosphorylation (Y694) of STAT5a could allow its dimerization, and phosphorylation (Y694) was a prerequisite for nucleus location. Some reports revealed that serine residues of STAT5a (S726 and S780, corresponding to murine S725 and S779) are phosphorylated in human myeloid malignancies including acute myeloid leukemia, and S779 phosphorylation, and the combined phosphorylation of S779 and S725 was necessary for the development of chronic myeloid leukemia, and sole S725 phosphorylation has no effect on disease development [10]. The combined effect of the more vital phosphorylation site Y694 with other phosphorylation sites such as S780 or S725 on cell proliferation, especially on DNA damage response, had not been reported in solid tumor.
In this report, we demonstrated the combined phosphorylation effect of Y694 with S780 on STAT5a nuclear location and target gene expression, and verified that IMB-HDC could intensively give rise to DNA breakage and cell apoptosis, and decreased STAT5a nuclear location, and give the first evidence to demonstrate that the combined decrease in Y694 and S780 phosphorylation was a prerequisite for the lessened STAT5a nuclear location treated by IMB-HDC in solid tumors, and that is the key for IMB-HDC anticancer effect exertion.
Materials and methods
Reagents and cells
Anti-γ-H2AX, anti-cleaved caspase3, anti-cleaved caspase9, anti-PARP, anti-p53, anti-RAD51, anti-phospho-STAT5A (Tyr 694), anti-STAT5A, and anti-c-myc antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-caspase2, anti-caspase8, and anti-caspase10 were purchased from Abcam (Cambridge, UK). Anti-phospho-STAT5A (Ser 726) and anti-phospho-STAT5A (Ser 780) were purchased from OriGene Technologies (Rockville, MD, USA). The anti-β-actin antibody was obtained from Sigma-Aldrich (Saint Louis, MO, USA), and peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies were purchased from ZSGQ-BIO Company (Beijing, China). Anti-γ-H2AX (Ser139), conjugated with fluorescein isothiocyanate (FITC), was purchased from BD Company (Franklin Lakes, NJ, USA). 7-Hydroxystaurosporine (UCN-01) and colcemid were purchased from Sigma-Aldrich (Saint Louis, MO, USA).
Cell lines
Human non-small-cell lung cancer cell A549, human pancreatic cancer cells ASPC-1 and MIA-Paca-2, human hepatoma cell HepG2, human breast cancer cell MCF-7, human colon carcinoma cells HCT-8 and HCT116, human breast cancer cell MDA-MB-231, human ovarian cancer cell SKOV3, human choriocarcinoma cell JEG3, human colon carcinoma cells HCT116 p53KO, and human leukemia cell K562 were obtained either from Cell Center of the Institute of Basic Medical Sciences or from ATCC. A549, ASPC-1, MIA-Paca-2, HepG2, MCF-7, HCT116, MDA-MB-231, HCT116 p53KO, and JEG3 cells were all cultured in the DMEM with 10% FBS at 5% CO2 at 37 °C. K562 cells was cultured in RPMI-1640 medium with 10% FBS. Anchorage-dependent cells in the exponential growth phase were harvested with a 0.25% trypsin–0.02% EDTA solution and resuspended in the specified medium. Suspension cells were subcultured regularly. Only single cells with viability over 95% (trypan blue exclusion) were used.
Cell growth inhibition assay
Cell growth inhibition was determined by using a SRB assay [11]. Cells were seeded in 96-well plates at 4 × 103–8 × 103/well and treated with the increasing concentrations of compound IMB-HDC and incubated for 24 h or 48 h, then cells were fixed with 50% trichloroacetic acid (TCA) (Sigma-Aldrich, Saint Louis, MO, USA), and then 0.4% (w/v) SRB in acetic acid (1%) was added. SRB-bound cells were solubilized with 10 mM Trizma base. The absorbance was read at 492 nm. The growth inhibition (%) was calculated at each concentration, and the IC50 was calculated by Sigmaplot. Assays were repeated three times, and the results were shown as mean and SD.
Colony-formation assay
Colony-formation assay was performed as described in ref. [12]. Briefly, HCT-8 or MIA-PaCa-2 cells were seeded in a six-well plate at 1000–2000/well, after 24 h escalating doses of IMB-HDC were added and continued to be incubated with cancer cells for 7 days, then cells were fixed with methanol for 15 min, and stained with 0.5% crystal violet for 15 min at room temperature. The colony is defined to consist of at least 50 cells. Visible colonies are counted. Colony-formation rate = (number of colonies/number of seeded cells) × 100%.
Comet assay
DNA breakage was measured with a neutral comet assay (Trevigen, Gaithersburg, MD, USA) as described in the manufacturer’s procedures and the literature. The treated cells were embedded in agarose on a slide and subjected to lysis followed by electrophoresis under neutral conditions. During electrophoresis, the damaged and fragmented negatively charged DNA migrated away from the nucleus toward the anode. The amount of migrated DNA was a measure of the extent of DNA damage. To detect DNA, the slides were stained with SYBR Gold (Life Technology, Inchinnan Business Park, Paisley, UK) staining solution. The slides were examined by fluorescence microscopy, and the results were analyzed with the comet analysis software CASP to quantify DNA damage [13]. For each drug concentration, three independent assays were conducted, in which comet tails were analyzed in a minimum of 50 randomly selected cells in each assay, and the parameter reflecting the DNA damage was represented as TL (tail length) [14].
Western blot
Whole-cell lysates were used for immunoblotting as described previously [12]. The cells were lysed using a lysis buffer (50 mmol/L Tris-HCl, pH 8.0, 150 mmol/L sodium chloride, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor). Twenty micrograms of protein lysate was resolved by SDS-PAGE and analyzed by immunoblotting with the specified antibodies (the dilution ratio of all primary antibodies was 1:1000, and the secondary antibody was 1:3000; usually 3 μL of the above-mentioned antibodies were used in the assay). The immunoreactive signals were revealed using the enhanced chemiluminescence (Millipore, Billerica, MA, USA) method and visualized with a ChemiImager 5500 imaging system (Alpha Innotech, San Leandro, CA, USA).
Immunofluorescence
γ-H2AX was detected by immunofluorescent microscopy as described in ref. [15]. Cells were treated with different concentrations of IMB-HDC and fixed in 4% paraformaldehyde, and then permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, Saint Louis, MO, USA) in PBS containing 5% BSA. Cells were incubated with γ-H2AX (phospho-Ser139) antibody FITC-conjugated at 30-fold dilution in 5% BSA in PBS overnight at 4 °C and mounted with mounting medium Vectashield (Vector Laboratories, Orton Southgate Peterborough, UK) and sealed with nail polish. γ-H2AX and nuclear staining were viewed with a Olympus FV500 Confocal Laser Scanning Microscope.
Analysis of cell cycle distribution
Flow cytometric analysis was performed as described in our previous report [12]. Cells were treated with IMB-HDC and collected at various time points; then they were harvested and fixed with 75% ethanol at −20 °C overnight. Cells were stained with propidium iodide (25 mg/mL) and RNase A (200 mg/mL) at 37 °C for 30 min. The DNA content was analyzed with a FACScan flow cytometer (COULTEREPICS XL, Fullerton, CA, USA).
Cell apoptosis assay
Quality analysis of apoptotic cells was accomplished with Annexin V-FITC Apoptosis Detection Kit (Beyotime, Haimen, Jiangsu, China) as described previously [16, 17]. Briefly, cells were treated with increasing concentrations of IMB-HDC, and after 48 h cells were collected for flow cytometry analysis by FACScan flow cytometry.
Cytogenetic analysis
Cytogenetic analysis was performed as described previously [15]. Briefly, colcemid (100 ng/mL) was added to cell cultures 2 h prior to harvest. Cells were collected and fixed in fresh methanol/acetic acid (3:1, v/v) at room temperature. After three washes with ice-cold methanol/acetic acid (3:1, v/v), cells were spread onto slides and air-dried overnight to stain with DAPI, and at least 50 metaphase spreads were analyzed on three separate slides for each sample.
Semiquantitative real-time PCR
RNA was extrated from untreated and IMB-HDC-treated cells expressing wild-type or mutant STAT5 variants using Trizol reagent (Invitrogen, Waltham, MA, USA). cDNA synthesis was performed with Oligo (dT) or random primers by using a Quantscript RT Kit (Tiangen). Real-time PCR was performed with SsoAdvanced SYBR GreenSupermix (Bio-Rad) and primers for Bcl-2 [10] and c-myc (forward 5′-AACAGGAACTATGACCTCG-3′, and reverse 5′-AGCAGCTCGAATTTCTTC-3′), CyclinD1 (forward 5′-ACAAACAGATCATCCGCAAACAC-3′ and reverse 5′- TGTTGGGGCTCCTCAGGTTC-3′), Pim-1 (forward 5′-TCTTCTGGCAGGTGCTG-3′ and reverse 5′-GGTAGCGAATCCACTCTG-3′), Osm (forward 5′-AGATACCTGAGCCCACACAGACAG-3′ and reverse 5′-ATCGTCCCATTCCCTGAAGACC-3′), and c-Fos (forward 5′-CGAAGGGAACGGAATAAGATGG-3′ and reverse 5′-AGACCTCCAGTCAAATCCAGGG-3′). Primers for target gene expression of STAT5 were normalized to GAPDH.
Site-directed mutagenesis
Site-directed mutagenesis constructs of eYPF-SAT5A (Y694E), eYPF-SAT5A (S780D), eYPF-SAT5A (Y694E + S780D), eYPF-SAT5A (Y694E + S780A), pCMV6-STAT5A-DDK (Y694E), pCMV6-STAT5A-DDK (S780D), pCMV6-STAT5A-DDK (Y694E + S780D), and pCMV6-STAT5A-DDK (Y694E + S780A) are finished using the QuikChange site-directed Mutagenesis Kit from Stratagene (La Jolla, CA, USA) according to the manufacturer’s instructions.
Transfection and establishment of cell lines stably expressing STAT5a and its mutant
Cells were collected and seeded in a six-well plate with 20 × 104 cells, and transfected with 2-μg constructs according to the manufacturer’s instructions with lipo2000 (Invitrogen, Waltham, MA, USA). Post transfection of pCMV6-STAT5a-flag or its mutants, cells were exposed to 800 µg/mL G418 (Invitrogen, Waltham, MA, USA) and screened for stable STAT5a and its mutant-expressing cells. (YFP)-tagged STAT5A and mutant’s protein were examined by immunofluorescent laser-scanning microscopy (Olympus IX71, 40-fold magnification).
In vivo antitumor activity
The in vivo efficacy of IMB-HDC was evaluated with HepG2 xenografts in nude mouse. Nude mice were purchased from Laboratory of Experimental Animals, Chinese Academy of Medical Sciences & Peking Union Medical College, and all animal studies were approved by the ethical board of Cancer Hospital Chinese Academy of Medical Sciences. In total, 1 × 107 HepG2 cells or cells stably expressing STAT5a or its mutants were suspended in 200 μL of PBS and inoculated s.c. in the right armpits of nude mice. After 3 weeks, the tumors were removed from the nude mice and dissected aseptically in sterile saline. Pieces of tumor tissue (3 mm3 in size) were then transplanted into the right armpits of nude mice with a trocar. Tumor-bearing mice were randomly divided into four groups (n = 5), when the tumor size was ~70 mm3. IMB-HDC (5, 10, or 20 mg/kg) was administered by intraperitoneal injection every second day until the mice were killed. Tumor size was measured every 3 days, and tumor volume was determined as length × width2/2. Mice were killed when the tumor volumes of the control group reached 1000 mm3; the tumor loads and tissues were isolated and used in further assays.
H&E staining and detection of apoptosis of tissue sections
Tumor and various tissues from IMB-HDC-administered mice were excised and preserved in liquid nitrogen until frozen sections were produced. H&E staining was performed as described in ref. [18]. Tumor tissues were prepared as homogenates to detect the corresponding proteins by Western blot.
Quantification and statistical analysis
Results were expressed as Mean±SD and analyzed using either two-tailed Student's t test or 2-way analysis of variance (ANOVA) in Graphpad Prism 5.0 software to assess statistical significance. P values <0.05 were considered to be statistically significant.
Results
IMB-HDC suppresses cancer cells proliferation in a time- and dose-dependent manner
To evaluate the effect of IMB-HDC on tumor cells, 5 μM IMB-HDC was added into the human colon carcinoma cell HCT-8, and cell morphology was observed at 3, 7, and 24 h. The cells were shrunk at 3 h and the spread of some cells was restricted; up to 7 h almost all cells could not spread. When the cells were treated for up to 24 h, some broken cells and cell debris were observed (Fig. 1b). After 48 h of treatment, most HCT-8 cells disappeared, and large amounts of cell debris were visible (data not shown), indicating a potent anticancer activity of IMB-HDC.
To evaluate the dose-dependent effect of IMB-HDC, 0 μM, 0.3 μM, 1.5 μM, 3.0 μM, and 6.0 μM IMB-HDC were added into the human colon carcinoma cell HCT-8, human leukemia cell K562, and human hepatoma carcinoma cell HepG2, and cell vitality was detected after incubation for 48 h. The results showed that IMB-HDC significantly inhibited cell growth in a dose-dependent manner (Fig. 1c). At the concentration of 0.3 μM, cell survival rate was ~95% of control and decreased to 70% at 1.5 μM after IMB-HDC treatment for 48 h in K562 cells (Fig. 1c, left); when the concentration was up to 3 μM, the survival rate was only 30% of control, indicating an obvious dose-dependent manner. Correspondingly, we also observed similar dose-dependent effects of IMB-HDC in HCT-8 cells and HepG2 cells (Fig. 1c, medium and right). Meanwhile, we observed the time-dependent effect with 3 μM IMB-HDC at 24 h, 48 h, and 72 h (Fig. 1d, left), and at 24 h the vital cell counts in HCT-8 cells were 55%, and up to 48 h and 72 h the viable cells decreased to 16% and 11%. In MIA-PaCa-2 cells treated at 3 μM, at 24 h, 48 h, and 72 h the viable cells were, respectively, 53%, 8%, and 3% of the control (Fig. 1d, medium), and HepG2 also got the corresponding time-dependent effect (Fig. 1d, right).
Given that colony-formation analysis is the gold standard of cytotoxic agent effect, we utilized colony-formation assay to further assess the inhibitory effect of IMB-HDC. HCT-8 cells or MIA-PaCa-2 cells were seeded in a six-well plate at 1000–2000 cells/well and incubated with escalating concentrations of IMB-HDC (0, 0.37, 0.75, 1.5, and 3 μM) for 7 days prior to colony count. The results turned out that IMB-HDC could inhibit colony formation of tumor cells in a dose-dependent manner. At 0.37 μM, colony-formation rate was ~90% of control and decreased to 60% at 0.75 μM, and at 1.5 μM the colony rate was only 20% of control and 5% at 3 μM, suggesting an obvious dose-dependent inhibitory effect on HCT-8 cells (Fig. 1e). Correspondingly, we also observed similar effects in MIA-PaCa-2 cells following exposure to IMB-HDC (Fig. 1e).
In addition, we also detected the inhibitory effect of IMB-HDC on other cancer cells, and the results showed that IMB-HDC could exhibit a spectral anticancer effect on various cells, including lung cancer, ovarian cancer, chorionic carcinoma of uterine, hepatoma, and breast cancer and leukemia (Fig. 1f), indicating a widely anticancer activity. Correspondingly, we also detected the effect of sophoridinol (the parent of IMB-HDC) on these tumor cells, and found that there was almost no inhibitory effect at 80 μM on seven tumor cell lines (Fig. 1g), but at the same concentration three-ring structure scafflold derivate IMB-HDC exhibited over 90% inhibitory effect in all tumor cells. These results showed that IMB-HDC possessed a stronger anticancer effect compared with its parent sophoridinol.
IMB-HDC induces cell apoptosis via the mitochondria-mediated apoptotic pathway
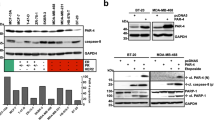
Cell apoptosis was assessed by flow cytometric analysis, and the results showed that IMB-HDC could extensively induce HCT-8 cells apoptosis in a dose-dependent manner, and at lower concentrations of 1 μM and 2 μM, the total apoptotic cells were 10.04% and 40% of the untreated cells, respectively, and at the higher concentration of 4 μM, the ratio was up to 65% (Fig. 2a), indicating that IMB-HDC plays its anticancer role in apoptosis inducement. Then we examined the corresponding apoptosis- signaling pathways, and the results turned out that IMB-HDC could lead to the final apoptotic signal protein caspase 3 and mitochondria-mediated apoptotic signal protein caspase 9 activation, but the signal proteins of non-mitochondrial-mediated apoptosis, such as caspase 2, caspase 8, and caspase 10, were not activated in HCT-8 and HepG2 cells (Fig. 2b). As the release of cytochrome c from the mitochondria to cytoplasm is essential in mitochondria-mediated apoptosis, we extracted the mitochondria and cytoplasm separately and detected the content of cytoplasmic cytochrome c, and the results suggested that cytochrome c was released to the cytoplasm, indicating that IMB-HDC-induced cancer cells apoptosis was mitochondrial-mediated (Fig. 2c).
a Indicated concentrations of IMB-HDC were added into HCT-8 cells for 48 h, and apoptosis was detected. b Various doses of IMB-HDC were added into HCT-8 and HepG2 cells, and the indicated proteins were examined. c IMB-HDC was added into HCT-8 cells, and the mitochondria and cytoplasm were extracted separately for cytochrome c detection.
IMB-HDC induces tumor cells DNA breakage, γ-H2AX foci formation, but does not activate p53 that is indispensable in DDR
Previous reports showed that IMB-HDC analog could induce DNA breakage [19, 20], and then we evaluated IMB-HDC effect on DNA breakage. Neutral single-cell gel electrophoresis was used to evaluate its effect on DNA breakage. The results showed that comet cells with comet tail were all visualized (Fig. 3a), indicating that IMB-HDC could induce cell DNA double-strand breakage. The quantitative data revealed that IMB-HDC caused the tail length increase in a dose-dependent manner in DNA breakage. At lower dose of IMB-HDC, the tail length was about 10-fold of control, and at the highest concentration of 8 μM, the tail length was up to 20-fold of control, indicating an extensive double DNA strand breakage.
a HepG2 cells were treated by increasing concentrations of IMB-HDC for 24 h, and alkaline comet electrophoresis assays were performed. The quantitative analysis was performed with the comet analysis software CASP, and the parameter TL (tail length) was employed to evaluate DNA damage. Assays were repeated three times, and the mean value of TL was normalized to the control (0 μM). b HepG2 cells were treated with elevating doses of IMB-HDC for 24 h or elevating treated time with 2 μM IMB-HDC (down); indicated proteins were detected by Western blot analysis. c HepG2 cells were treated with IMB-HDC for 24 h, γ-H2AX foci were tested with γ-H2AX antibody conjugated with FITC, the nucleaus was stained with DAPI, and cells were observed with a Confocal Laser Scanning Microscope. d HepG2 cells were treated with IMB-HDC, and at the indicated time points cells were collected for cycle analysis. Mean±SD. *P < 0.05; ** P < 0.01. Scale bar represents 5 μm.
Given that the histone variant H2AX is phosphorylated rapidly on Ser139 (γ-H2AX) in response to DNA double-strand breakage in cells, and it is a maker of DNA breakage [21], we examined the γ-H2AX level and the results showed that the γ-H2AX expression significantly increased in the addition of IMB-HDC and its elevation was in a dose-dependent manner (Fig. 3b). Then we utilized immunofluorescent analysis to further examine whether γ-H2AX foci were formed (–H2AX foci represent a DNA damage site in the nucleus) [22] in the presence of IMB-HDC, and the results showed that IMB-HDC could cause the abundant formation of γ-H2AX foci in the nuclei of HCT-8 cells after 24 h of treatment (Fig. 3c). Meanwhile, we observed that IMB-HDC could induce G2/M-phase arrest (Fig. 3d). All the above results indicated that IMB-HDC could significantly lead to DNA breakage in a short time and these damages had been persistent in response to IMB-HDC.
In DNA damage response, acting as the pivotal linker to accept signals from upstream DDR sensors (such as ATR) to the downstream effector to exert DNA repair, generally p53 plays the indispensable role in sensors transmission. When p53 was activated by its upstream signal, p53 stability was enhanced and subsequently triggered its target gene expression to induce DNA repair, apoptosis, and cycle arrest [16, 17, 21, 23, 24]. In our study, although we observed DNA breakage and apoptosis following the addition of IMB-HDC, we did not observe p53 expression increase, and ATR activation was blocked that is different from the general DNA-damaging agents (Fig. 3b), suggesting that a novel DDR mechanism implicated in IMB-HDC induced DDR and apoptosis, and the induced apoptosis was independent of p53.
IMB-HDC induces chromosomal aberration
Based on the above-mentioned results, the DNA breakage was the dominant factor underlying anticancer activity of IMB-HDC. To observe DNA damage more intuitively, in line with the previous report [19], we performed cytogenetic analysis of HCT-8 cells treated with colcemid to force cells to stay in the metaphase. This could allow to observe chromosomal damage, as only in a metaphase chromosomes are relaxed and the damage could be easily observed [20, 22, 25]. UCN-01, a Chk1 kinase inhibitor, was used to abrogate IMB-HDC-induced G2 arrest [19]. UCN-01 could allow G2-arrested cells to proceed through mitosis and into the metaphase to observe the chromosome damage, and our results showed that 100 nM UCN-01 treatment alone had no discernible effects on cell cycle distribution (Fig. 4a). By scoring 50 well-resolved metaphases, we detected an average of four chromatid-type aberrations, including gaps and breaks, per cell (Fig. 4b). In contrast, fewer aberrations were visualized in untreated cells and UCN-01-treated cells alone (data not shown).
HCT-8 cells were treated with 3 μM IMB-HDC for 24 h, 100 nM UCN-01 was added, and culturing continued for 6 h to abrogate M-phase arrest prior to sample collection. Colcemid (100 ng/mL) was then added to the cell cultures 2 h prior to harvesting. a Representative images of chromosome spreads. Arrows, chromosome breaks. b Quantitation and comparison of chromosome aberrations. Mean±SD. *P< 0.01 vs Control or UNC-01 alone, # P= 0.1 vs Control. Scale bar represents 2.5 μm.
IMB-HDC suppresses the phosphorylation of Y694 and S780 in STAT5a and blocks nuclear translocation of STAT5a
Our previous chip results of sophoridine derivate (data not shown) whose structure was similar to IMB-HDC showed that the expression of STAT5a target genes decreased; meanwhile recent data showed that in DDR STAT5a is an essential molecule in contribution to DDR, and its silence contributed to anticancer effect of DNA-damaging therapy [9]. Thus, we speculated that STAT5a also might be involved in IMB-HDC-induced DNA damage and apoptosis. In DDR, p53 expression increased and then initiated STAT5a expression in response to DNA damage. Subsequently, STAT5a translocates into the nucleus to arouse Rad51 expression, it is involved in the search for homology and strand-pairing stages in homologous recombination of DNA breakage [26, 27] to activate ATR to promote broken DNA repair.
In our study, we observed that IMB-HDC only could induce minor expression change in STAT5a or p53 (Figs. 3b, 5a), but STAT5a target gene Rad51 obviously decreased (Fig. 5a); correspondingly we also detected the translational level of other STAT5a target genes associated with apoptosis and proliferation, such as CyclinD1, c-myc, bcl-2, c-fos, and Pim-1, and the results revealed that their mRNA levels all decreased (Fig. 5b). These data suggested that although STAT5a expression did not obviously change, its target gene expression associated with apoptosis and proliferation decreased significantly, hinting that there might be an unconventional mechanism involved in IMB-HDC anticancer mechanism.
a HepG2 cells were treated with 5 μM IMB-HDC at various time points, and indicated protein was detected with Western blot. b HepG2 cells were treated with increasing doses of IMB-HDC, RNA was extracted, and the mRNA level of indicated gene was examined with real-time PCR. c IMB-HDC was added into HepG2 cells, and the cytoplasm and nuclear protein were extracted separately for Western blot and indicated proteins were detected. d STAT5a-eGFP was transfected into HCT-8 cells prior to the addition of IMB-HDC, and cells were observed with immunofluorescence microscopy. e HepG2 cells were treated with IMB-HDC for 24 h, STAT5a antibody was added into cells prior to the addition of a secondary antibody conjugated with FITC, nucleus was stained with DAPI, and cells were observed with a Confocal Laser Scanning Microscope. Mean±SD. *P < 0.05, **P< 0.01 vs Control. Scale bar represents 5 μm.
Acting as a transcriptional factor, STAT5a activity exertion needs nuclear location. Considering that the total STAT5a slightly decreased and expression of its target gene decreased significantly, thus we hypothesized that there might be the less nuclear STAT5a to induce the expression decrease of its target gene, then we examined nuclear STAT5a content, and found that the content of nuclear STAT5a significantly decreased by 70% of control, and the nuclear–cytoplasmic ratio of STAT5a was lowered down by 80%, indicating an obvious movement from the cytoplasm to the nucleus, and IMB-HDC could suppress STAT5a nuclear translocation (Fig. 5c). Then we used the construct eGFP-STAT5a to transfect HCT-8 cells, and detected IMB-HDC-mediated nuclear location effect with the newly expressed STAT5a. The results demonstrated that IMB-HDC really potentiated newly synthesized STAT5A to remain in the cytoplasm (Fig. 5d). Furthermore, we utilized confocal assay to assess the STAT5a location with STAT5A antibody, and found that nuclear STAT5a was almost not observed after IMB-HDC treatment (Fig. 5e). These results revealed that IMB-HDC could lessen STAT5a nuclear translocation.
Since the total STAT5a level did not change and nuclear STAT5a decreased obviously, we speculated that there might be a movement from the cytoplasm to nucleus. Tyrosine phosphorylation (Y694) of STAT5a is known to be a vital factor to promote STAT5a nuclear translocation by inducement of STAT5a dimer formation, and most attention has been paid to Y694 [18]. But recent reports showed that STAT5a serine phosphorylation (S726 and S780) has been implicated in STAT5a nuclear location to promote leukemogenesis, and S725 and S780 phosphorylation is a prerequisite during BM transplantation. Then we detected the phosphorylation of Y694, S726, and 780 in the addition of IMB-HDC in HCT-8 cells. The results showed that IMB-HDC could significantly depress the phosphorylation of Y694 and S780 in a time-dependent manner, and up to 6 h their phosphorylation was not nearly visualized (Fig. 5a), but the level of 726 phosphorylation did not change up to 12 h (Fig. 5a).
IMB-HDC depresses STAT5a nuclear translocation, transcriptional activity, and triggers DNA breakage and apoptosis via blocking 694 and 780 phosphorylation
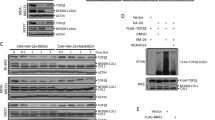
Then we wanted to verify whether lessened phosphorylation of 694 and 780 of STAT5A plays the role in IMB-HDC-induced nuclear translocation, subsequent transcriptional activity, DNA breakage, and apoptosis, and thus constructed the consecutive activation mutant Y694E, in which Y694 was mutated to glutamic acid (E) and was consecutively phosphated to lead to STAT5a location in the nucleus [28]. Meanwhile according to the above-mentioned results, we constructed the mutant S780D in which site S780 was mutated to aspartic acid (D) and was consecutively phosphorylated, and the mutants Y694E + S780D in which Y694 and S780 were both consecutively phosphated, and Y694E + S780A in which Y694 was consecutively phosphated but S780 was not phosphated. The verification of these mutations is shown in Fig. 6a (down).
HCT-8 cells were transfected with STAT5a-eGFP or its mutants prior to the addition of IMB-HDC, and cells vitality was examined with SRB assay, and indicated proteins were detected with Western blotting (a). STAT5a location was observed with immunofluorescence microscopy (b), mRNA levels of STAT5a target gene were detected with real-time PCR (c), and apoptosis-associated or indicated proteins were detected with Western blot (d). Comet assay to examine DNA breakage was assayed (e), and DNA damage proteins were detected (f). Mean±SD. *P<0.05 vs Y694E.
The results showed that sole Y694E and Y694E + S780D all could rescue IMB-HDC-induced growth inhibition, indicating that sole 694 or both of Y694 and S780 might be the targets of IMB-HDC. To further validate whether S780 is also the target of IMB-HDC, we further compared the effect of between Y694E + S780A (Y694 was consecutively phosphated, but S780 was not phosphated) and Y694E + S780D in addition of IMB-HDC; the results showed that Y694E + S780A did not reverse IMB-HDC-induced growth inhibition effect, indicating that S780 is essential for IMB-HDC effect and S780 is also the target of IMB-HDC.
Correspondingly, we also examined the effect of decreased phosphorylation of S780 and Y694 on nuclear translocation of STAT5a in addition of IMB-HDC, and subsequent transcription level of STAT5a target gene by transfecting eGFP-STAT5a and its mutants. Similar to the above-mentioned assays, we observed that 694 consecutive phosphorylation alone could rescue IMB-HDC-induced lessened nuclear STAT5a (Fig. 6b). In Y694E + S780A-transfected cells, we also observed that Y694E (694 consecutive phosphorylation) that induced the enhancement of STAT5a nuclear location was rescued by 780 inactivation in addition of IMB-HDC, indicating that the combined decrease in 694 and 780 phosphorylation was vital to IMB-HDC-induced shortened nuclear location of STAT5A. These results mentioned above further demonstrated that decreased phosphorylation of 780 is also required for the function exertion of our compound in addition to decreased phosphorylation of 694.
Then, we assayed the transcriptional level of target gene of STAT5a associated with proliferation, and the results showed that its transcriptional levels were consistent with STAT5a nuclear location in Fig. 6b (Fig. 6c), indicating that decreased phosphorylation of 694 and 780 was necessary for IMB-HDC anticancer effect. Although sole 694 also could play a role in IMB-HDC-induced STAT5a nuclear location and growth inhibition, these effects depend on 780 decreased phosphorylation, and sole 780 did not affect IMB-HDC activity. The apoptosis data and Rad51 changes also supported the above-mentioned results (Fig. 6d).
Then we utilized comet assays to detect the DNA breakage and got the similar results that 694 consecutive activation could rescue IMB-HDC-induced DNA breakage, but this rescue could be reversed by 780 inactive mutation, indicating that combined decrease of 780 and 694 phosphorylation was essential to IMB-HDC-mediated DNA breakage (Fig. 6e). γ-H2AX, the represent protein marker of DNA breakage, was examined and the corresponding results were observed, and the changes also support the comet data of the mutants (Fig. 6f). All the above-mentioned results demonstrated that IMB-HDC depressed STAT5a nuclear translocation, transcriptional activity, and triggers DNA breakage and apoptosis via blocking 694 and 780 phosphorylation
IMB-HDC-induced proliferation inhibition depends on the decreased phosphorylation of 694 and 780 in vivo
Next, in a tumor xenograft nude mouse model, we examined IMB-HDC anticancer efficacy. The results indicated that IMB-HDC could suppress tumor xenograft proliferation, and at 10 mg/kg the inhibitory rate was ~61%, while at 2.5 mg/kg the inhibitory rate was ~37% (Fig. 7a). The body weight curves indicated that the animals tolerated well the IMB-HDC dosages administered (Fig. 7b). When the tumor sizes reached 1000 mm3, the mice were killed, and tumors and various tissues were excised to be used for further analysis. Tissues were prepared as sections for H&E staining, and tumor tissues were used for WB analysis. The results showed that IMB-HDC could significantly induce cleaved PARP and reduced phosphorylation of 694 and 780, indicating that IMB-HDC could induce apoptosis and inhibit 694 and 780 phosphorylation in vivo (Fig. 7c). H&E staining of various tissues showed that there were no obvious pathologic changes observed between control and administrative groups (Fig. 7d). Then we inoculated stable Y694E or Y694E + S780A-expressing cells in the right armpits of nude mice, and when the tumor volume reached 80 mm3, IMB-HDC was administered every second day, and tumor volume was measured. The in vivo results were in line with cell results, that is, when S780 was mutated into alanine and not be phosphorylated, the promoting proliferation activity of Y694E (694 consecutive phosphorylation) was eliminated, and phosphorylation of both 694 and 780 was required for IMB-HDC anticancer activity induction (Fig. 7e).
Nude mice (n = 5) bearing HepG2 xenografts were administered IMB-HDC i.p. × 1 every second day after tumor implantation. The results were recorded every 3 days after tumor implantation with all mice alive. Mice were killed when the tumor volume of the control group reached 1000 mm3; the tumor loads and other tissues were isolated and prepared sections. a Tumor volume was measured by calipers twice per week in the indicated days. Mean±SD. **P<0.01. b Body weights of mice-harboring tumors were monitored twice per week in the indicated days. c Western blot for analysis of p-STAT5a and apoptotic-associated proteins from various treatment groups. d H&E staining of tissue sections from various treatment groups. e Stable Y694E–STAT5a and Y694E + S780A–STAT5a-expressing cells were inoculated in the armpits of nude mice, and when the tumor volume reached 80 mm3, IMB-HDC was administered for 21 days once per day, and tumor volume was measured by calipers twice per week in the indicated days, and the relative volume to STAT5a was calibrated and the volume-treated day curve was depicted. Scale bar represents 20 μm.
Discussion
The Chinese traditional medicine Fufang Kushen injection was approved as an anticancer agent in 1995, and the main active ingredient of Kushen extract is sophoridine and it exhibits the anticancer effect. Based on the original sophoridine structure, our lab made a series of optimization, modification, and synthetized the corresponding sophoridine derivatives, and found that IMB-HDC possessed the potent anticancer activity in various tumor cell lines in vitro and in vivo. H&E staining of various tissues disclosed that there was no obvious toxicity, indicating the safety of IMB-HDC (Fig. 8).
Although previous studies revealed that sophoridine derivatives could induce DNA breakage, apoptosis, and inhibit DNA repair, further studies about how to trigger apoptosis and inhibit DNA repair did not elucidate clearly. Our studies found that p53, the pivotal linker to accept signals from upstream DDR sensor and transmit these signals to the downstream effector to exert DNA repair or apoptosis, could not be activated and its expression did not change, indicating that there might be a novel mechanism involved in IMB-HDC-induced DNA damage response. Our previous chip assay analysis showed that the level of several STAT5a target genes decreased; thus, we speculated that STAT5a might be implicated in IMB-HDC-induced apoptosis and DNA breakage in tumor cells.
STAT5a expression has been studied closely to tumor, such as prostate cancer, colorectal cancer, and head and neck cancer [25,26,27]. Aberrant continual activity of STAT5 has been causally linked to human leukemias and solid tumor formation [28]. Unphosphorylated or inactive STAT5a may suppress tumor growth in colorectal cancer, and active STAT5a expression in premalignant and tumor lesions has shown potential as a prognostic marker in oral squamous cell carcinoma [26, 29]. Inhibition of STAT5a/b has resulted in large-scale apoptotic death [27].
STAT5a classically can sense cytokine and growth factor signals in the cytoplasm and deliver those signals to responsive genes in the nucleus to regulate DNA damage, proliferation, differentiation, growth, and apoptosis [28]. Movement between cytoplasmic and nuclear compartments is thereby central to their biological function. Although in our study we did not observe that STAT5a expression decreased and that is consistent with p53 unchanged expression as the target gene of p53, we found that after IMB-HDC treatment nuclear STAT5a content was obviously lowered and cytoplasm STAT5a increased, indicating that IMB-HDC might promote STAT5a nuclear translocation via regulating its movement between the cytoplasm and nucleus. Correspondingly, we also observed the expression of STAT5a target genes associated with apoptosis and genes beneficial to DNA repair gene Rad51 eliminated. Rad51 is an enhancing DNA repair protein and its elimination will attribute to DNA breakage, and that might a contributor to IMB-HDC-induced DNA damage. Other target genes of STAT5a such as Bcl-2, c-myc, and CyclinD1 decrease would contribute to growth inhibition and apoptosis, and these changes might be involved in IMB-HDC-induced anticancer effect on tumor cells.
Although tyrosine phosphorylation (Y694) of STAT5a is known to be beneficial for the dimer formation that is prerequisite to STAT5a shuttle from the cytoplasm to nucleus, recent reports showed that phosphorylation of S726 and S780 was also implicated in the STAT5a shuttle. But the combined effect of Y694 and other two serines on STAT5a nuclear location was not further reported in solid tumors. In our report, we visualized that IMB-HDC inhibits significantly the phosphorylation of Y694 and S780, and had a little effect on the phosphorylation of S726, indicating that the decreased phosphorylation of Y694 and S780 might be implicated in lessened STAT5a nuclear location. Our further study revealed that consecutive phosphorylation at Y694 alone could reverse IMB-HDC-mediated lessened STAT5a nuclear location, DNA breakage, and growth inhibition, but this rescue could be inhibited by S780 consecutive unphosphorylation, indicating that although Y694 alone could play its role, its function still depends on S780 phosphorylation. Simultaneous phosphorylation decrease of Y694 and S780 is indispensable to IMB-HDC anticancer effects on tumor cells, indicating a novel mechanism of phosphorylation of both Y694 and S780 on STAT5a nuclear location.
Although for decades the anticancer study of sophoridine and its derivate has been going on, the molecular mechanism of how to induce apoptosis and DNA breakage has not been clarified. Our results for the first time showed that IMB-HDC could inhibit the phosphorylation of STAT5a Y694 and S780, and subsequently block the shuttle of STAT5a from the cytoplasm to nucleus, and then decreased the expression of STAT5a target gene associated with apoptosis and proliferation to exert anticancer activity. Meanwhile STAT5a target gene RAD51 expression also decreased and subsequently inhibited ATR activation. Furthermore, we first verified that although phosphorylation of STAT5a Y694 is indispensable to STAT5a nuclear shuttle, this effect also depends on S780 phosphorylated state (unchanged or elevated phosphorylation), and S780 also plays a key role in STAT5a nuclear shuttle. This mechanistic exploration is extremely vital to the combination of drug therapy, such as the inhibitory effect of IMB-HDC on STAT5a 694 and 780 phosphorylation, hinting that IMB-HDC could be used in combination with other kinase inhibitors, and the inhibitory effect of IMB-HDC on DDR provides the probability that IMB-HDC could be used in combination with DNA-damaging agents. Regarding the further mechanism of IMB-HDC to block the phosphorylation of STAT5a, we speculated that it might be a result that IMB-HDC hampers STAT5a phosphorylation, either IMB-HDC inhibits STAT5a kinase enzyme activity or hinders the binding of STAT5a with its kinase, and in the future we will continue to investigate them.
IMB-HDC possesses good safety profiles, good druggable characteristics, high solubility, the special chemical scaffold, and good PK profile in vivo [6, 29], suggesting that it is a promising agent and worthy to be explored further [31].
References
Nie HCJ, Gao YQ, Jin SG, Wang LT. The anti-hbv study of sophoridine in vitro. J Beijing Univ Tradit Chin Med. 2007;26:678–80.
Yang ZZY, Cao XQ. The effect of sopho ra alopecuroides on cv b3 in vitro. J Sichuan Univ Med Sci. 2003;21:14–16.
Lu HSY, Cao M, Song XW, Tang Y, Yang YJ. Effects of sophoridine on cardiac function and myocardial ultrastructure of rats with myocardial infarction. J Second Milit Med Univ. 2012;33:470–3.
Li XWY, Chen SL. The effect of sophoridine on tumor cells. Acta Pharmacol Sin. 1987;8:153–8.
Li X-m, Wu Y-g, Pan D-x, Wu L, Yu Y, Zhang A-h, et al. Sophoridine is a new antitumor medicine with new molecular structure. J N. Drugs. 2006;15:654.
Bi C, Ye C, Li Y, Zhao W, Shao R, Song D. Synthesis and biological evaluation of 12-n-p-chlorobenzyl sophoridinol derivatives as a novel family of anticancer agents. Acta Pharm Sin B. 2016;6:222–8.
Zhao W, Zhang C, Bi C, Ye C, Song D, Liu X, et al. Sophoridinol derivative 05d induces tumor cells apoptosis by topoisomerase1-mediated DNA breakage. Onco Targets Ther. 2016;9:2805–17.
Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62.
Maranto C, Udhane V, Hoang DT, Gu L, Alexeev V, Malas K, et al. Stat5a/b blockade sensitizes prostate cancer to radiation through inhibition of rad51 and DNA repair. Clin Cancer Res. 2018;24:1917–31.
Berger A, Hoelbl-Kovacic A, Bourgeais J, Hoefling L, Warsch W, Grundschober E, et al. Pak-dependent stat5 serine phosphorylation is required for bcr-abl-induced leukemogenesis. Leukemia. 2014;28:629–41.
Zhao W, He H, Ren K, Zhang H, Chen Y, Shao R. Myofibrillogenesis regulator-1 promotes cell adhesion and migration in human hepatoma cells. Chin Sci Bull. 2013;58:3007–14.
Zhao W, He H, Ren K, Li B, Zhang H, Lin Y, et al. Mr-1 blocks the megakaryocytic differentiation and transition of cml from chronic phase to blast crisis through mek dephosphorylation. Blood Cancer J. 2013;3:e107.
Zhao W, Jiang G, Bi C, Li Y, Liu J, Ye C, et al. The dual topoisomerase inhibitor a35 preferentially and specially targets topoisomerase 2alpha by enhancing pre-strand and post-strand cleavage and inhibiting DNA religation. Oncotarget. 2015;6:37871–94.
Rojas E, Lopez MC, Valverde M. Single cell gel electrophoresis assay: methodology and applications. J Chromatogr B Biomed Sci Appl. 1999;722:225–54.
Zhao W, Liu H, Wang J, Wang M, Shao R. Cyclizing-berberine a35 induces g2/m arrest and apoptosis by activating yap phosphorylation (ser127). J Exp Clin Cancer Res. 2018;37:98.
Taylor WR, Stark GR. Regulation of the G2/m transition by p53. Oncogene. 2001;20:1803–15.
Williams AB, Schumacher B. P53 in the DNA-damage-repair process. Cold Spring Harb Perspect Med. 2016;6:a026070.
Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb Protoc. 2008;2008:pdb prot4986.
Guo L, Liu X, Nishikawa K, Plunkett W. Inhibition of topoisomerase iialpha and g2 cell cycle arrest by nk314, a novel benzo[c]phenanthridine currently in clinical trials. Mol Cancer Ther. 2007;6:1501–8.
Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, et al. The chk1 protein kinase and the cdc25c regulatory pathways are targets of the anticancer agent ucn-01. J Biol Chem. 2000;275:5600–5.
Ravizza R, Gariboldi MB, Passarelli L, Monti E. Role of the p53/p21 system in the response of human colon carcinoma cells to doxorubicin. BMC Cancer. 2004;4:92.
Sampath D, Cortes J, Estrov Z, Du M, Shi Z, Andreeff M, et al. Pharmacodynamics of cytarabine alone and in combination with 7-hydroxystaurosporine (ucn-01) in aml blasts in vitro and during a clinical trial. Blood. 2006;107:2517–24.
Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–55.
Pauklin S, Kristjuhan A, Maimets T, Jaks V. Arf and atm/atr cooperate in p53-mediated apoptosis upon oncogenic stress. Biochem Biophys Res Commun. 2005;334:386–94.
Hotte SJ, Oza A, Winquist EW, Moore M, Chen EX, Brown S, et al. Phase i trial of ucn-01 in combination with topotecan in patients with advanced solid cancers: a princess margaret hospital phase ii consortium study. Ann Oncol. 2006;17:334–40.
Holloman WK. Unraveling the mechanism of brca2 in homologous recombination. Nat Struct Mol Biol. 2011;18:748–54.
Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, et al. Rad52 inactivation is synthetically lethal with brca2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–91.
Si Y, Ji X, Cao X, Dai X, Xu L, Zhao H, et al. Src inhibits the hippo tumor suppressor pathway through tyrosine phosphorylation of lats1. Cancer Res. 2017;77:4868–80.
Bi C, Zhang C, Li Y, Tang S, Wang S, Shao R, et al. Synthesis and biological evaluation of sophoridinol derivatives as a novel family of potential anticancer agents. ACS Med Chem Lett. 2014;5:1225–9.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFA0201504), National Natural Science Foundation of China (Nos. 81473249 and 81102464), the National Mega-project for Innovative Drugs (2014ZX09201042), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-2-002), and Drug Innovation Major Project of China (2018ZX09711001-007-002).
Author information
Authors and Affiliations
Contributions
WLZ, DQS, and RGS contributed to the experimental design; WLZ, YX, MYW, RGS, and other authors contributed to the acquisition and analysis of the data; RGS reviewed the paper; WLZ and RGS obtained the funding; WLZ wrote the paper. WLZ, RGS, and DQS contributed to the experimental design; WLZ, YX, YHQ, CY, YL, XJL, MYW, and CWB contributed to the acquisition and analysis of the data; RGS reviewed the paper; WLZ and RGS obtained the funding; WLZ wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All mentioned authors were involved in the writing of this paper and gave consent for its publication.
Rights and permissions
About this article
Cite this article
Zhao, Wl., Xing, Y., Ye, C. et al. The novel quinolizidine derivate IMB-HDC inhibits STAT5a phosphorylation at 694 and 780 and promotes DNA breakage and cell apoptosis via blocking STAT5a nuclear translocation. Acta Pharmacol Sin 41, 686–697 (2020). https://doi.org/10.1038/s41401-019-0333-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0333-6