Abstract
Study design
Method development.
Objectives
To develop a reliable protocol for automatic segmentation of Thoracolumbar spinal cord using MRI based on K-means clustering algorithm in 3D images.
Setting
University-based laboratory, Tehran, Iran.
Methods
T2 structural volumes acquired from the spinal cord of 20 uninjured volunteers on a 3T MR scanner. We proposed an automatic method for spinal cord segmentation based on the K-means clustering algorithm in 3D images and compare our results with two available segmentation methods (PropSeg, DeepSeg) implemented in the Spinal Cord Toolbox. Dice and Hausdorff were used to compare the results of our method (K-Seg) with the manual segmentation, PropSeg, and DeepSeg.
Results
The accuracy of our automatic segmentation method for T2-weighted images was significantly better or similar to the SCT methods, in terms of 3D DC (p < 0.001). The 3D DCs were respectively (0.81 ± 0.04) and Hausdorff Distance (12.3 ± 2.48) by the K-Seg method in contrary to other SCT methods for T2-weighted images.
Conclusions
The output with similar protocols showed that K-Seg results match the manual segmentation better than the other methods especially on the thoracolumbar levels in the spinal cord due to the low image contrast as a result of poor SNR in these areas.
Similar content being viewed by others
Introduction
The spinal cord is a tubular structure of the central nervous system located in the vertebral column and surrounded by bony columns and soft tissues extending from the medulla oblongata. The spinal cord is undeniably involved in many functions of the nervous system, and its magnetic resonance imaging (MRI) in health and disease became very interesting for clinicians and researchers. For example, spinal cord injury (SCI) is one of the primary causes of motor disabilities in humans and Skeletal muscles experience deleterious physiological changes after an accident [1, 2]. Due to the highly variable nature of recovery following an injury to the cord, it is difficult to predict the outcome and prognosis [3, 4]. It is important to have an accurate assessment of injury severity in SCI as early as possible to plan the acute injury management and have a better idea about the prognosis of the patient that helps the procedures in later stages, clinical trials, and candidacy for novel therapies [4, 5]. However, due to the existing challenges such as brain trauma and pain, an objective assessment seems to be far to reach [4, 5]. Advances in the MR imaging promises new opportunities for the study of spinal cord injuries and other conditions and is becoming the standard technique for the assessment of the damage [4,5,6,7]. However, to increase the impact there is a need for the improvement of techniques that may contribute to the validity and reliability of measures. Since SC segmentation is the first step in atlas-based SC analysis [7], improvement of this step can have a drastic influence on the outcome of the analysis. Manual segmentation techniques are time-consuming and subject to between and within rater variability [7, 8]. Accordingly, for large sample sizes and clinical purposes, manual segmentation is a vulnerability. The Spinal Cord Toolbox has contributed invaluably to the development of protocols for the automatic segmentation of the SC [8] but most of their focus was on the cervical cord in the human and lumbar cord has not received proper attention. Besides, the algorithm used in their protocol is more sensitive to coexisting pathologies that may influence the outcome of the segmentation and its accuracy. A segmentation protocol that can deal with these challenges in the SCI is required to improve the impact of MR imaging on the study of SCI by contributing to the validity and reliability of measures. Similarly, in other clinical populations like low back pain patients and patients with Multiple Sclerosis (MS) improved outcome of segmentation the lumbar cord can contribute to the use of MR imaging more accurately in the clinic. It has been shown that atrophy in the gray matter of the spinal cord is associated with disability in patients with MS [9, 10]. So, it is important for clinicians and researchers dealing with those patients to monitor the structural changes in the cord (its shape or its cross-sectional area (CSA)). Hence, understanding the pathophysiological sequelae would help to prevent and reduce disease burden and would facilitate the development of effective regenerative and neuroprotective treatments. However, manual segmentation of the cord is time-consuming, unreliable and varies from person to person [11]. Raters need to segment each scan in parallel and for each subject, the associated consensus segmentation of the raters for the cord and canal must be estimated using majority voting. For this purpose, the dice coefficient (DC) and Hausdorff distance (HD) between each rater’s segmentation usually is calculated and a consensus mask is produced across all the raters’ marks as a gold standard. To overcome these issues, a segmentation model is required to find the severity of the injury and to predict the disease patterns along the segmented spinal cord regions. Automatic detection of spinal cord and calculation of the CSA metrics (the rate of volumetric changes and tissue atrophy) are complex due to changes in structure and size. Besides automatic segmentation of this area is an essential factor that influences the detection of spinal cord atrophy and its severity of the SCI. Although semi or fully automated methods are susceptible to the level of contrast to noise in the image, they use sophisticated methods that lead to robust outcomes and more reliable results which indeed can contribute to the reproducibility of studies. For example, among those hired semi-automated methods Tench et al. [12] improved this metric in their edge-detection based method by taking into account the spinal cord orientation and the partial volume effect between spinal cord and cerebra-spinal fluid (CSF). However, these methods need more manual interventions (requires a few points along the spinal cord, identified by the user to initialize the segmentation process). Other researchers developed techniques based on an active surface model [13, 14], used a double threshold-based method on the 3D T2-weighted turbo spin-echo MR scans of the spinal cord [15], proposed a protocol based on a globally minimal path optimization method using PCA to cluster the spinal cord shapes [16], or developed a method based on one-dimensional template matching [17]. A significant limitation of all these methods goes back to them requiring the intervention of the user at different stages, which may influence the reliability at different levels. On the other hand, fully automated methods are preferred because they are faster, suitable for bigger samples and not susceptible to the user’s bias. For example, De Leener et al. [8, 18] developed an automatic segmentation method (PropSeg) based on multi-resolution propagating of tubular deformation models on MR images. Consequently, Gros et al. [11] proposed an original and fully automatic framework (DeepSeg) based on convolutional neural networks (CNNs) applied to the spinal cord morphometry for segmenting the spinal cord and/or intramedullary MS lesions, degenerative cervical myelopathy (DCM), neuromyelitis optica (NMO), traumatic SCI, amyotrophic lateral sclerosis (ALS), and syringomyelia (SYR) from a variety of MRI contrasts and resolutions. However, none of these methods are optimized from images acquired from the thoracolumbar spinal cord. Due to the specific structure of this part of the spinal cord and its involvement in damages related to lower limbs and lower back, it is important to have a reliable protocol for the segmentation of the cord and the canal.
In this paper, we present a fully automatic framework for the segmentation of the spinal cord and spinal canal, optimized for thoracolumbar segments. The main contributions of this paper are: (1) providing an independent detection module to find the spinal cord and spinal canal location based on the circular shape. The symmetry of the body helps to perform Circular Hough Transform to find the circular shape better. (2) Applying an anisotropic diffusion (AD) filter to remove noises and stabilize the optimization process on the results of images. (3) Using the K-means clustering algorithm for segmentation of the spinal cord and spinal canal. Our method performs well in low SNR regions, and it is robust towards low contrast, especially in thoracolumbar areas. Since this technique is optimized for low contrast images, we predict a better match between manual segmentation and K-Seg output as compared with two established models implemented in the SCT.
Methods
Image acquisition
The method described in this paper was tested on T2-weighted MRI images of the thoracolumbar spinal cord of 20 uninjured volunteers (male, age = 24.48 ± 4.62 years, range = 22–31 years). The imaging data were acquired on a 3T Siemens Prisma MR scanner at the National Brain Mapping Laboratory, the University of Tehran. Volunteers were positioned carefully, and pads were used to restrict foot and spine movements. A structural volume was acquired in the sagittal orientation using a T2 sampling perfection by using flip angle evolution (SPACE) sequence (TR = 1500 ms; TE = 119 ms; FOVs = 320 × 320 mm; matrix size = 256 × 256 × 56 slices, slice thickness = 1.3 mm, and in-plane resolution = 1 × 1 mm). To demonstrate the efficiency of the K-Seg in the segmentation of the cord in the low contrast region, only the vertebrae below T7 were included. The K-Seg was implemented in MATLAB environment. The code and sample data are freely available at (see Supplementary Appendix 2 for MATLAB Code).
Segmentation framework
The K-Seg framework is illustrated in Fig. 1. It consists of five major steps:
- (1)
recognition of region of interest (ROI) based on mutual information (MI) as a similarity measure in left–right direction
- (2)
applying a canny filter to extract the edges and Hough line transform to remove the extra parts from the ROI in the anterior–posterior direction
- (3)
using a Circular Hough Transform to find appropriate circles around the SC and CSF along the cord. So the automatic CSF segmentation can be achieved as well as SC location
- (4)
resorting the circles by selecting a candidate circle close to the AP line (a circle with radius [7,8,9] is good enough to find the spinal cord curvature in each slice due to the spine has varying shape from top to bottom)
- (5)
applying an AD filter to remove noises and segmentation of the spinal cord and canal by K-means clustering algorithm to classify intensities.
The framework includes: (1) recognition of region of interest (ROI) based on mutual information as a similarity measure; (2) applying canny filter to extract edges and Hough line transform in order to remove the extra parts from ROI in anterior–posterior direction; (3) computing the centerline of spinal cord by using Hough circular transform; (4) resorting the circles by select a candidate circle close to AP line with radius [7,8,9] to find the spinal cord curvature and (5) applying an anisotropic diffusion filter to remove noises and segmentation of spinal cord and canal by k-means clustering algorithm to classify intensities.
Extracting region of interest in the left–right direction
Most methods rely on semiautomatic or manual approaches, getting one or more landmarks to detect the position of the ROI to extract the spine in images [13,14,15] and some groups do it automatically [8, 11, 18,19,20,21,22]; we used an automatic method: an axial slice was automatically selected (e.g., the middle slice of the volume). Using the MI metric [23] and presuming the human body as symmetric the medial anterior–posterior line (AP line) was detected (the AP line passes the spinal cord) (Fig. 1, step 1). This step, in particular, reduced the computational time. Detection of the AP line using the MI was through the following equation [18, 23]:
where S (Ileft, Iright) represents the MI between images, and Ileft and Iright are two 2D axial planes in left and right sides. A restrained image is built here by cropping an area with eight slices in the left–right direction around the AP line.
Removing unwanted regions inside of the ROI in the anterior–posterior direction
Here we aimed to remove unwanted regions inside of the ROI obtained from the previous step in the anterior–posterior direction (Fig. 1, step 2). To extract the spinal canal position, finding more details of the edges on the ROI is essential. Among the many edge-detection methods, we used the Canny method [24], because of its ability to detect more details of edges in an image. After the edge detection, the Hough Line Transform was used to find the vertical lines in the image to extract the edges of interest [25, 26].
The Hough transform is a technique to isolate features of particular shapes within an image. The most common use of the Hough transform is in the detection of curves, lines, circles, and ellipses. This method is robust and unaffected by the image noise. We assume that the Hough Lines Transform is parameterized in this form [25, 26]:
Where p is a perpendicular distance from the line to the origin, and θ is the angle of distance p from the x-axis.
The Hough line transform (for removing unwanted tissues inside of the ROI in the AP direction) is based on the following steps:
- (1)
Binarizing the ROI in the sagittal view, and applying the Hough line transform to it;
- (2)
Finding 50 significant Hough transform peaks (enough to indicate vertical lines of the spinal cord)
- (3)
Detecting the initial and endpoints of lines, and linking these points on the restrained binary image. Linking these points obtained the spinal cord and canal’s range in the image and let us remove the area outside these lines.
Detection of spinal cord and canal location
The detection module, based on the circular Hough transform, is in three steps (Fig. 1, step 3). A Circular Hough Transform [25, 26] was applied to the restrained image in the axial view (considering the circular shape of the spinal cord and the canal). Among the many identified circles, the circle with the minimum distance to the AP line was selected as a candidate circle (Fig. 1, step 4). The distance was measured by the Euclidean method. The circular Hough transform is parameterized in this form [25, 26]:
where (a, b) is the center of the circle and r is its radius. The radius of the circle of interest was in the range of 7–9 mm (as the spine has a varying shape from top to bottom), and the sensitivity was set on 0.97. Next, in each axial slice, we created a mask on the candidate circle and then assigned the gray level intensities from the original image to this circle (Fig. 1, step 4). By estimating the center of the circle in each slice, the coordinate of the centerline could be continuously updated in each slice to estimate the cord’s curvature. In the slices where no optimal circle could be identified (e.g., due to the low contrast of the image), the algorithm used the coordinates of the circle of the previous slice. As the candidate circles include spinal cord and canal tissues as well, the K-means clustering algorithm was applied to classify the intensities and segment the spinal cord and CSF regions (Fig. 1, step 5). This clustering method is sensitive to the image noise, and therefore, the images were first spatially smoothed. To avoid the blurring of the edges while smoothing, the contrast of the image was enhanced by applying an AD filter (Fig. 1, step 5).
Anisotropic diffusion filter
Diffusion algorithms by Perona–Malik formulation remove noise from an image using a partial differential equation. The isotropic diffusion equation was:
Where I (x, y, 0) is the original image, (x, y) refers to the spatial position, t is an artificial time parameter and ∇I is the image gradient [27, 28]. Because of equivalency of the isotropic diffusion with a Gaussian filter, Perona and Malik [28] replaced the classical formula of an isotropic equation with:
Where \(\left| {\left| {\nabla {\mathrm{I}}} \right|} \right|\) is the gradient magnitude of the image and \(({\mathrm{g}}\left| {\left| {\nabla {\mathrm{I}}} \right|} \right|)\) is an “edge-stopping” function. This function is defined to satisfy g(x)→0 when x→∞. So that the diffusion is stopped across the edges. This filter can smooth the original image without any edge blurring by preserving brightness discontinuities.
Spinal cord and canal segmentation by K-means clustering algorithm
K-means is a powerful, simple, and fast clustering algorithm [29, 30]. A clustering method is used to divide a set of data points into several groups (Fig. 1, step 5). The K-means algorithm on an image operates in these steps:
- (1)
initializing the number of clusters (k = 3);
- (2)
calculating the Euclidean distance d between the center of the clusters and each pixel of an image, using the following equation [29, 30]:
$${\mathrm{d}} \,=\, \left| {\left| {{\mathrm{ p}}\left( {{\mathrm{x}},{\mathrm{y}}} \right) \,-\, {\mathrm{c}}_{\mathrm{k}}} \right|} \right|$$(6) - (3)
Assigning each pixel to the nearest center in a cluster based on distance d and recalculating the new position of the center using this equation:
$${\mathrm{c}}_{\mathrm{k}} \,=\, \frac{1}{{\mathrm{k}}}\mathop {\sum }\limits_{{\mathrm{y}} \in {\mathrm{ck}}} \mathop {\sum}\limits_{{\mathrm{x}} \in {\mathrm{ck}}} {{\mathrm{p}}({\mathrm{x}},{\mathrm{y}})}$$(7) - (4)
Repeating the process until it satisfies the tolerance or error value;
- (5)
Reshaping the cluster pixels into the image.
The number of clusters that we consider to segment spinal cord and canal areas is (k = 3). The first cluster is related to the spinal cord region, the second class includes the spinal canal area and the third cluster is referred to keep the same false areas which added on both SC and CSF throughout the detecting circles. By switching among these clusters the automatic SC and CSF segmentation can be achieved as well.
Inter-rater variability of the spinal cord segmentation
We estimated the inter-rater variability of the spinal cord segmentation between two raters on all volunteers (n = 20). For each of these subjects, one scan was available, which allow the raters to segment each scan in parallel and combine their information at the end of the work. For this purpose, we calculated the DC and HD between each rater’s segmentation and a consensus mask produced as majority voting across all the raters’ marks for a gold standard (see Table 1) [11].
Validation methodology
The segmented data by K-Seg was validated against (1) manual segmentation performed independently by two experienced individuals and also the consensus of two expert manual segmentations which was selected as a gold standard, (2) segmentation using the PropSeg [8, 18], implemented in C++ based on multiresolution propagation of tubular deformation models, and (3) segmentation using the DeepSeg [11], implemented in Python 2.7 based on CNNs. To assess the performance of the K-Seg, two measures were computed as below:
- (1)
The 3D DC defined in [31] by the following equation:
$${\mathrm{Dice}}\left( {{\mathrm{X}},{\mathrm{Y}}} \right) \,=\, \frac{{2\left| {{\mathrm{X}} \cap {\mathrm{Y}}} \right|}}{{\left| {\mathrm{X}} \right| \,+\, \left| {\mathrm{Y}} \right|}},$$(8)where X, Y are the binary segmentation mask to compare. As stated, the consensus of two expert manual segmentations was considered as a gold standard, and the three methods were compared with that. The DC range is between [0, 1], and closing to 1 means more similarity to the gold standard.
- (2)
The HD [32] that is described as the maximum distance between two images. Two sets are close in the HD if every point of either set is close to some points of the other set. A low HD demonstrates good results in comparison.
$${\mathrm{H}}\left({\mathrm{X}},{\mathrm{Y}}\right) \,=\, \max \left\{{\mathrm{h}}\left({\mathrm{X}},\,{\mathrm{Y}} \right),{\mathrm{h}}\left({\mathrm{Y}},\,{\mathrm{X}} \right)\right\},$$(9)where h (X, Y) is the HD from the surface X to Y and is defined as:
$${\mathrm{h}}\left({\mathrm{X}}, {\mathrm{Y}} \right) = \mathop{\mathrm{max}}\limits_{{\mathrm{x}}\epsilon {\mathrm{X}}}\left\{\mathop{\mathrm{min}}\limits_{{\mathrm{y}}\epsilon {\mathrm{Y}}} \left\{{\mathrm{d}}\left({\mathrm{x}},\, {\mathrm{y}} \right) \right\} \right\},$$(10)where x, y are two points from the surfaces X, Y and d (x, y) is the Euclidean distance between a and b.
Two independent one-way ANOVAs [33] with DC and HD as the dependent variables and the segmentation method versus manual segmentation as the independent variable compared the outcome of the K-Seg versus DeepSeg and PropSeg.
Results
Results of the spinal cord segmentation on T2-weighted images on 20 uninjured subjects are presented in Table 1. High DC and low HD demonstrate good results of the K-Seg method. In addition, we calculated the DC and HD between each rater’s segmentation and a consensus mask produced across all the raters’ labels as a gold standard. As shown in Table 1, results have been compared with two independent experienced individuals and also the consensus of two expert manual segmentations as a gold standard. The accuracy of the proposed method was found to be close, comparable and in some situations better than a single rater, in terms of 3D DC (p < 0.001). The 3D DCs were respectively 0.81 and HD 12.3 by the K-Seg method for T2-weighted images. Therefore, K-Seg shows the accurate result when compared with the SCT methods, as shown by the higher DC and lower HD in Table 1. Two examples of K-Seg segmentation of the spinal cord are presented in Fig. 2a, b. Our findings suggest better results by using the K-Seg as compared with both DeepSeg and PropSeg. These images have a loss of quality with distortion in the presence of magnetic material. So the proposed method manages the segmentation firmly even when CSF and SC contrast is at lowest on a specific part of the spinal cord. As illustrated in Fig. 2a, b, some segmentation errors can be observed by PropSeg and DeepSeg methods, particularly in the lumbar portions. Furthermore, for a typical T2-W acquisition (TR = 1500 ms; TE = 119 ms; FOVs = 320 × 320 mm; matrix size = 256 × 256 × 56 slices, slice thickness = 1.3 mm, in-plane resolution = 1 × 1 mm), the computation time on a workstation with WIN 10 OS system equipped with an (Intel core i7, 2.20 GHz processor and 6 GB RAM), was 40 s for K-Seg versus 1 min 55 s for DeepSeg and 32 s for PropSeg.
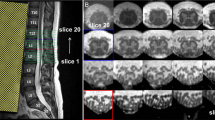
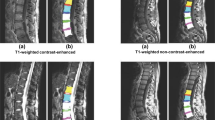
a Example of automatic spinal cord segmentation. The framework includes segmentation on T2-W MRI data in the sagittal view. This is a comparison between the original image from left to right with manual (yellow), PropSeg (green), DeepSeg (blue), and K-Seg(red). The dice coeficient indicated below each method. b Example of spinal cord segmentation in axial and sagittal views. The framework includes segmentation in axial and sagittal views, each color corresponds to one method related to vertebrae level (lower slice→L2, middle slice→T12, upper slice→T10), the red color is related to K-Seg, green and the blue color correspond to DeepSeg and PropSeg segmentation.
Cross-sectional areas
Spinal cord measurements such as the CSA can be extracted by K-Seg framework. CSAs were calculated for each slice of a binarized segmentation of the spinal cord. This measurement was calculated for three methods by counting the pixels in each slice and then comparing it with a gold standard. The differences in CSA observed at vertebrae levels in Fig. 3. Significant differences were observed for multiple vertebral levels. The CSAs in T12 level are greater than other vertebral levels.
Mean and standard error of the mean (SEM) extracted from T2-W images are plotted on the same scale by corresponding vertebral levels. Significant differences are visible among methods results. Each color corresponds CSA to one method, the yellow color is presented by the manual segmentation of spinal cord, the red color is related to K-Seg, green and the blue color corresponds to DeepSeg and PropSeg segmentation. Also, each axial slice corresponds to one vertebrae level (Left slice→L2, middle slice→T12, Right slice→T10) with manual segmentation.
Discussion
In this paper, we described the development and validation of a framework for the automatic segmentation of the spinal cord and spinal canal. This framework also allows us to calculate the quantitative CSA metric along the spinal cord for multiple vertebral levels. As stated earlier, segmentation of the spinal cord, spinal canal and also calculation of the quantitative CSA metric along the spinal cord became very interesting for clinicians and researchers. So segmentation frameworks will develop for automated SC segmentation on subjects without any pathological damage besides patients with a range of spinal pathologies, including those with traumatic SCI [1,2,3,4,5] and MS [9, 10]. In this cohort, we extended the method (k-seg) by providing segmentation of both SC and CSF based on the concept of the K-means clustering algorithm. We demonstrated the ability of k-seg to accurately segment the SC and CSF on T2-W images based on the same initialization. The first step of the method initializes by using a Circular Hough Transform to find an appropriate circle around the SC and CSF along the cord. The second phase of the method is applying an AD filter to remove noises and stabilize the optimization process on the curvature which was detected in the previous section. Finally, the k-means clustering algorithm is used to separate SC and CSF regions from each other. Also, the sensitivity of the location of initialization for spinal cord segmentation along the axial plane is important. So to initialize the algorithm, thoracic regions of the spine (e.g., the middle slices of the selected volumes) yielded better results because of the higher contrast and shape of the spinal cord. Therefore, detecting the spinal cord position (the first candidate circle) in these areas is more reliable than the other parts [18]. Based on this, we concluded that the initialization of our method in the middle axial slices of the MR image would obtain better results.
As we mentioned earlier, fully automated methods are preferred because they are faster, suitable for bigger samples and not susceptible to the user’s bias. For example, Koh et al. [22] proposed a 2D active contour on sagittal T2-W MRI scans using gradient vector flow. Neubert et al. [20] proposed an automated 3D segmentation method on vertebral bodies and intervertebral discs from MRI based on statistical shape analysis and template matching of gray level intensity profiles. De Leener et al. [8, 18, 19] developed an automatic segmentation method (PropSeg) based on multiresolution propagating of tubular deformation models on MR images. Finally, Gros et al. [11] proposed an original and fully automatic framework (DeepSeg) based on CNNs applied to the spinal cord morphometry for segmenting the spinal cord and/or intramedullary MS lesions, DCM, NMO, traumatic SCI, ALS, and SYR from a variety of MRI contrasts and resolutions. Although many of the abovementioned methods, and in particular the PropSeg and DeepSeg, perform the segmentation of spinal cord from images with different contrasts and fields of view very well, when it comes to the segmentation of the thoracolumbar cord as compared with the cervical and thoracic cord, these tools do not work as well since in thoracolumbar cord as compared with the cervical cord the SNR is much lower and variation in shape and length of the cord inside the vertebral column is higher [11, 18]. The segmentation process is highly dependent on the quality of images and works better on images with high contrast between SC and CSF regions. In images with a lower CNR both manual and automatic segmentation face a lot of difficulties. We hypothesized that k-seg outperforms the existing protocols for the automatic segmentation of the spinal in the regions with higher noise and lower SNR and CNR. We were able to compare the outcome of our segmentation protocol with other protocols in different slices across the thoracolumbar spinal cord. Interestingly, we could see that as we move to lower slices in the lumbar cord, the gap between the performance of k-seg, deepseg and propseg tends to enlarge and we can see more errors in the results acquired from deepseg and propseg (Table 1). Besides, as shown in Supplementary Appendix 1 for Supplementary Figures, by moving from thoracic segments to the lumbar spinal cord, the level of the noise increases significantly and SNR decreases consequently. These findings support our hypothesis on the better performance of k-seg than deepseg and propseg when it comes to the segmentation of images with a higher level of noise. Indeed, a better performance by k-seg when it comes to the segmentation of the spinal cord in areas with a higher level of noise can be attributed to the utilization of a well-established denoising filter (AD Filter). Applying a proper filter on MR images is worth being taken into account because most of the smoothing filters can suppress important details along with the spinal cord segmentation, such as edges and small scale atrophy. However, AD filter allows us to combine the two most important attributes of the denoising algorithm: edge preservation and noise removal. A comparison between selected filters in our method and other methods in SCT depicts that the propseg method uses adaptive contrast properties are included within the deformable model framework that appropriately deal with potential lack of signal-to-noise ratio [19]. However, this deformable framework only applies a Gaussian filter through the model which blurs the image and can hinder the edge-detection process. Also, no smoothing filter was mentioned in deepseg method. The motivation behind the use of the AD filter in k-seg is to overcome the blurring effects of the Gaussian smoothing approach. In this approach, the image is only convolved in the direction orthogonal to the gradient of the image, which ensures the preservation of edges. Taking this step often requires to remove image artifacts beforehand to make k-means clustering more robust and efficient to segment SC/CSF intensities. So, we showed that the k-seg algorithm could follow the shape of the spinal cord even when the cord and CSF contrast is minimal on a significant portion of the spinal cord.
Limitations
Despite the good precision of K-Seg in T2-W thoracolumbar 3D images especially in the lower part of the spine, which has poor contrast, our segmentation method failed in particular occasions (mostly in the initializing step by detecting a non-target circle among many circles, using the circular Hough transform method). Also, our segmentation framework is sensitive to the quality of the images, which could be partly overcome by choosing a suitable filter to remove noises.
In addition, the selection of T2-W images as an MR imaging protocol is considered in the present study because the quality of T1W images in the thoracolumbar region is very low and many groups are not interested in doing so. Accordingly, in most databases, only T2-W images are included. Similarly, in our database, we only had access to T2-W images and could not get access to images with T1w contrasts. However, we could test the algorithm on diffusion images from two subjects tested on another scanner than ours, and the outcome was similar to what we could get for T2-W images. Since the number of subjects was not enough for statistical analysis, we decided not to present them in the current paper.
Conclusion
The current study was aimed to present a fully automatic segmentation method supporting T2-weighted images which can work efficiently on the thoracolumbar levels in the spinal cord. We also compare the outcome of the segmentation using the K-Seg with the outcome of manual segmentation and existing widely used methods (i.e., PropSeg and DeepSeg). The output with similar protocols showed that K-Seg results match the manual segmentation better than the other methods especially on the thoracolumbar levels in the spinal cord. Future works are needed to replicate these results on spine images in a different field of view.
Data archiving
All the data for this study are available upon request submitted to the corresponding author.
References
Seif M, Gandini Wheeler-Kingshott CA, Cohen-Adad J, Flanders AE, Freund P. Guidelines for the conduct of clinical trials in spinal cord injury: neuroimaging biomarkers. Spinal Cord. 2019;57:717–28.
Freund P, Seif M, Weiskopf N, Friston K, Fehlings MG, Thompson AJ, et al. MRI in traumatic spinal cord injury: from clinical assessment to neuroimaging biomarkers. Lancet Neurol. 2019;18:1123–35.
Ahuja CS, Wilson JR, Nori S, Kotter MRN, Druschel C, Curt A, et al. Traumatic spinal cord injury. Nat Rev Dis Prim. 2017;3:1–21. https://doi.org/10.1038/nrdp.2017.18.
Talekar K, Poplawski M, Hegde R, Cox M, Flanders A. Imaging of spinal cord injury: acute cervical spinal cord injury, cervical spondylotic myelopathy, and cord herniation. Semin Ultrasound, CT MRI. 2016;37:431–47.
Bozzo A, Marcoux J, Radhakrishna M, Pelletier J, Goulet B. The role of magnetic resonance imaging in the management of acute spinal cord injury. J Neurotrauma. 2011;28:1401–11.
Fehlings MG, Martin AR, Tetreault LA, Aarabi B, Anderson P, Arnold PM, et al. A clinical practice guideline for the management of patients with acute spinal cord injury: recommendations on the role of baseline magnetic resonance imaging in clinical decision making and outcome prediction. Glob Spine J. 2017;7:221S–30S.
Lévy S, Guertin M, Khatibi A, Mezer A, Martinu K, Chen J, et al. Test-retest reliability of myelin imaging in the human spinal cord: Measurement errors versus region- and aging-induced variations. PLOS ONE. 2018;13:e0199796. 1–25 https://doi.org/10.1371/journal.pone.0189944.
De Leener B, Kadoury S, Cohen-Adad J. Robust, accurate and fast automatic segmentation of the spinal cord. Neuroimage. 2014;98:528–36. https://doi.org/10.1016/j.neuroimage.2014.04.051.
Schlaeger R, Papinutto N, Panara V, Bevan C, Lobach IV, Bucci M, et al. Spinal cord gray matter atrophy correlates with multiple sclerosis disability. Ann Neurol. 2014;76:568–80. https://doi.org/10.1002/ana.24241.
Lin X. Spinal cord atrophy and disability in multiple sclerosis over four years: application of a reproducible automated technique in monitoring disease progression in a cohort of the interferon -1a (Rebif) treatment trial. J Neurol Neurosurg Psychiatry. 2003;74:1090–4.
Gros C, De Leener B, Badji A, Maranzano J, Eden D, Dupont SM, et al. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. 2018. http://arxiv.org/abs/1805.06349.
Tench CR, Morgan PS, Constantinescu CS. Measurement of cervical spinal cord cross-sectional area by MRI using edge detection and partial volume correction. J Magn Reson Imaging. 2005;21:197–203.
Horsfield MA, Sala S, Neema M, Absinta M, Bakshi A, Sormani MP, et al. Rapid semi-automatic segmentation of the spinal cord from magnetic resonance images: Application in multiple sclerosis. Neuroimage. 2010;50:446–55.
Coulon O, Hickman SJ, Parker GJ, Barker GJ, Miller DH, Arridge SR. Quantification of spinal cord atrophy from magnetic resonance images via a B-spline active surface model. Magn Reson Med. 2002;47:1176–85.
El Mendili M-M, Chen R, Tiret B, Villard N, Trunet S, Pélégrini-Issac M, et al. Fast and accurate semi-automated segmentation method of spinal cord MR images at 3T applied to the construction of a cervical spinal cord template. PLoS ONE. 2015. https://doi.org/10.1371/journal.pone.0122224.
Kawahara J, McIntosh C, Tam R, Hamarneh G. Globally optimal spinal cord segmentation using a minimal path in high dimensions. IEEE 10th International Symposium on Biomedical Imaging: From Nano to Macro, ISBI. IEEE Computer Society, 2013; pp. 848–851. https://doi.org/10.1109/ISBI.2013.6556608.
Cadotte A, Cadotte DW, Livne M, Cohen-Adad J, Fleet D, Mikulis D, et al. Spinal cord segmentation by one dimensional normalized template matching: a novel, quantitative technique to analyze advanced magnetic resonance imaging data. PLoS ONE. 2015;10:e0139323. https://doi.org/10.1371/journal.pone.0139323.
De Leener B, Cohen-Adad J, Kadoury S. Automatic segmentation of the spinal cord and spinal canal coupled with vertebral labeling. IEEE Trans Med Imaging. 2015;34:1705–18.
De Leener B, Taso M, Cohen-Adad J, Callot V. Segmentation of the human spinal cord. Magn Reson Mater Physics. Biol Med. 2016;29:125–53. http://www.ncbi.nlm.nih.gov/pubmed/26724926.
Neubert A, Fripp J, Shen K, Salvado O, Schwarz R, Lauer L, et al. Automated 3D segmentation of vertebral bodies and intervertebral discs from MRI. In Proceedings of IEEE DICTA. 2011; pp. 19–24.
Koh J, Scott PD, Chaudhary V, Dhillon G. An automatic segmentation method of the spinal canal from clinical MR images based on an attention model and an active contour model. In: 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; 2011. p. 1467–71. http://ieeexplore.ieee.org/document/5872677/.
Koh J, Kim T, Chaudhary V, Dhillon G. Automatic segmentation of the spinal cord and the dural sac in lumbar MR images using gradient vector flow field. Conf Proc IEEE Eng Med Biol Soc. 2010;3117–20.
Unser M, Thevenaz P. Optimization of mutual information for multiresolution image registration. IEEE Trans Image Process. 2000;9:2083–99. http://ieeexplore.ieee.org/document/887976/.
Canny J A. Computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell. 1986;PAMI-8:679–98. http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=4767851.
Ballard DH. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recognit. 1981;13:111–22. https://www.sciencedirect.com/science/article/abs/pii/0031320381900091.
Perasso A, Campi C, Massone AM, Beltrametti MC. Spinal Canal and Spinal Marrow Segmentation by Means of the Hough Transform of Special Classes of Curves. In: Murino V., Puppo E. (eds) Image Analysis and Processing. ICIAP 2015. Lecture Notes in Computer Science, vol 9279. Springer, Cham.
Black MJ, Sapiro G, Marimont DH, Heeger D. Robust {A}nisotropic {D}iffusion. IEEE Trans Image Process. 1998;7:421–32.
Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12:629–39. http://ieeexplore.ieee.org/document/56205/.
Patel PM, Shah BN, Shah V. Image segmentation using K-mean clustering for finding tumor in medical application. Int J Comput Trends Technol. 2013;4:1239–42. http://www.ijcttjournal.org.
Dhanachandra N, Manglem K, Chanu YJ. Image segmentation using K-means clustering algorithm and subtractive clustering algorithm. Procedia Comput Sci. 2015;54:764–71. https://doi.org/10.1016/j.procs.2015.06.090.
Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. https://doi.org/10.2307/1932409. http://doi.wiley.com/10.2307/1932409.
Aspert N, Santa-Cruz D, Ebrahimi T. MESH: Measuring Errors between Surfaces using the Hausdorff Distance. In IEEE Multimedia. 2002;705–8.
Kim H-Y Analysis of variance (ANOVA) comparing means of more than two groups. Restor Dent Endod. 2014;39:74–7. http://www.ncbi.nlm.nih.gov/pubmed/24516834.
Author information
Authors and Affiliations
Contributions
SS, SAB, and MAO were involved in study design, method development, and manuscript preparation. HD and AK were involved in study design, method development, data acquisition, and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
We certify that all applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during the course of this research.
Informed consent
All the data were anonymized before the processing and participants gave written consent for sharing their data.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sabaghian, S., Dehghani, H., Batouli, S.A.H. et al. Fully automatic 3D segmentation of the thoracolumbar spinal cord and the vertebral canal from T2-weighted MRI using K-means clustering algorithm. Spinal Cord 58, 811–820 (2020). https://doi.org/10.1038/s41393-020-0429-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41393-020-0429-3
This article is cited by
-
Magnetic resonance image segmentation of the compressed spinal cord in patients with degenerative cervical myelopathy using convolutional neural networks
International Journal of Computer Assisted Radiology and Surgery (2022)
-
Nanoscopic subcellular imaging enabled by ion beam tomography
Nature Communications (2021)