Abstract
BACKGROUND
Tetralogy of Fallot (TF) is a severe congenital defect of heart development. Fine-tuned sequential activation of Notch signaling genes is responsible for proper heart chamber development. Mutations in Notch genes have been associated with TF. The aim of this study was to analyze the activity of the Notch pathway in cardiac mesenchymal cells derived from ventricular tissue of TF patients.
METHODS
Cardiac mesenchymal cells were isolated from 42 TF patients and from 14 patients with ventricular septal defects (VSDs), used as a comparison group. The Notch pathway was analyzed by estimating the expression of Notch-related genes by qPCR. Differentiation and proliferation capacity of the cells was estimated.
RESULTS
The TF-derived cells demonstrated a dysregulated pattern of Notch-related gene expression comparing to VSD-derived cells. Correlation of Notch signaling activation level by HEY1/HES1 expression level with proliferation and cardiogenic-like differentiation of cardiac mesenchymal cells was observed but not with clinical parameters nor with the age of the patients.
CONCLUSIONS
The data suggest a contribution of dysregulated Notch signaling to the pathogenesis of tetralogy of Fallot and importance of Notch signaling level for the functional state of cardiac mesenchymal cells, which could be critical considering these cells for potential cell therapy approaches.
Similar content being viewed by others
Introduction
Congenital heart disease (CHD) is the most common type of birth defect, affecting approximately 8/1000 live births in all countries. Tetralogy of Fallot (TF) is the most common complex, cyanotic CHD, with a prevalence of 1/3000 births.1 The morphological phenotype is characterized by four malformations: narrowing of the right outflow tract (pulmonary stenosis), ventricular septal defect (VSD), dextraposition of the aorta to the right side over the VSD (an overriding aorta), and hypertrophy of the right ventricle (RV). TF is a classical cyanotic CHD caused by a subpulmonary obstruction with shunting mainly from the right to the left ventricle via the VSD leading to an ejection of poorly oxygenated blood into the aorta. At present, surgical repair is performed where the stenosis of the right outflow tract is relieved and the VSD is closed, enabling exclusive ejection of oxygenated blood via the left ventricle.2 TF patients have signs of cardiomyocyte hypertrophy and disarray, varying degrees and types of fibrosis, and cardiomyocyte degeneration that are considered to be a consequence of high RV pressure and hypoxemia.2
The cause of TF is still not known and no single candidate gene is associated with the disease phenotype. Approximately 20% of cases are associated with a recognized syndrome or chromosomal anomaly.3 Most significantly, approximately 15% of TF patients have 22q11.2 deletion syndrome, wherein the major causal gene is TBX1.4 In approximately 80% of TF cases that are non-syndromic, there is generally no identifiable cause, largely due to their non-Mendelian pattern of inheritance.5 Accordingly, a polygenic genetic architecture has been hypothesized and genome-wide approaches have been undertaken to provide insights into the complex genetic alterations responsible for TF and other CHDs.2
Whole-exome sequencing has been used successfully to identify new CHD candidate genes. Given the complex nature of cardiac development, it is not surprising that multiple transcription factors and signaling molecules responsible for cardiogenesis have been implicated in TF, with well-characterized genes GATA4, NKX2.5, FOXC2, TBX5, and TBX1.6 It is assumed that disruption of the balanced interaction of transcription factors important to cardiogenesis plays a significant role in non-syndromic TF.
Mutations in NOTCH1, NOTCH2, and JAG1 genes of the Notch family have also been associated with TF, but more commonly, mutations in these genes produce other heart malformations.7 Nevertheless, there are several lines of evidence that Notch genes play a role in TF. Copy number variants altering NOTCH1 and JAG1 have been identified in TF patients.8 In another study, RV tissues of TF patients were analyzed, and the expression of the majority of genes associated with the Wnt and Notch pathways was significantly reduced.9 A very recent study shows that NOTCH1 locus is a frequent site of genetic variants predisposing to non-syndromic TF, with 6% of patients exhibiting rare, deleterious variants.10
Notch (1–4) are single-pass transmembrane receptors that play a critical role in cell fate decisions.11 Sequential expression of Notch components and related genes is indispensable during development of the heart and vessels.12 In the heart, the Notch pathway is required for regulation of key pathways involved in cardiac trabeculae formation, and sequential activation of several Notch genes supports myocardial patterning, maturation, and compaction.12 As Notch signaling is particularly important for cardiovascular development, mutations in genes encoding Notch signaling components lead to various types of cardiac and vascular disorders. Moreover, we and others have recently shown that a wider spectrum of cardiovascular diseases related to left ventricular outflow tract development is associated with dysregulation of the whole Notch pathway even without obvious mutations in Notch-related genes.13,14,15,16,17
Taking into account the importance of Notch signaling in cardiac development and patterning, we hypothesized that the Notch pathway might be attenuated in the cells of heart tissue of TF patients, and this dysregulation might be responsible for the remodeling of cardiac tissue of TF patients. Cardiac progenitor or mesenchymal cells deriving from ventricular and atrial tissues are widely described for human and rodent hearts.18 Although the reparative potential of these cells remains questionable, they represent a convenient and relatively accessible source of human cardiac cells and could be used to uncover mechanisms of processes related to cardiac development.19,20 The biology and the role of these cells in the heart are not yet fully understood. The aim of this study was to analyze the activity of the Notch pathway in cardiac mesenchymal cells (CMCs) derived from ventricular tissue of TF patients. As a comparison group, we used CMCs from the tissues of patients with isolated VSDs, as this isolated defect does not show Notch involvement.21 Our data suggest a contribution of the dysregulation of Notch genes in cardiac cells to the pathology of TF and confirm that fine-tuned Notch signaling is one of the key factors responsible for proper heart development.
Methods
Patients
The clinical research protocol was approved by the local Ethics Committee of the Almazov National Medical Research Center and was in accordance with the principle of the Declaration of Helsinki. All patients’ representatives gave informed consent.
Samples were harvested during surgical intervention at the Almazov National Medical Research Center. Patient data are presented in Table 1. Patients with genetic syndromes were excluded from the study. During correction of VSD, fragments of the right atria (RAs) were harvested (n = 14). During repair of TF, fragments of the RV were harvested (n = 42).
Primary cultures
CMCs from myocardial tissue were isolated, propagated, and differentiated as described previously.22 Briefly, myocardial tissue was dissected onto small clamps and digested by collagenase II (Worthington) solution (1 mg/mL) for 2 h at 37 °C. The cell suspension was then passed through a 40-µm strainer, centrifuged at 300 × g for 5 min, and cells were resuspended in growth medium (22.5% endothelial cell growth medium-2 (Stem Cell), 67.5% M199, 10% fetal bovine serum (Hyclone), 1× nonessential amino acids, 50 units/mL penicillin, and 50 µg/mL streptomycin (Thermo Fisher Scientific)), seeded on 0.1% gelatine-coated flask, and expanded. After expansion on a 25-cm2 flask, the cells were reseeded (passage 1) and maintained in Petri dishes (Corning) in growth medium at 37 °C and 5% CO2. For each patient, one CMC line representing a mixed population derived from myocardial tissue was analyzed. All the lines were analyzed at the third passage.
Human umbilical vein endothelial cells (HUVECs) were used as a control lacking cardiogenic properties. The cells were harvested from umbilical vein by enzymatic dissociation as described.16 The cells were cultured in Petri dishes covered with 0.2% gelatin in phosphate-buffered saline (ScienCell).
The immunophenotype of CMCs (data not shown) was evaluated by flow cytometric analysis performed on CytoFlex (Beckman Coulter). Cells were resuspended in 100 µL of phosphate-buffered saline containing 1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO), incubated for 20 min at 20 °C in the dark with the following monoclonal antibodies (Abs): anti-CD146 phycoerythrin (PE; Beckman Coulter, A07483), anti-CD166 PE (Beckman Coulter, A22361), anti-PDGFRβ allophycocyanin (APC; BD Pharmingen, FAB1263A), anti-CD31 PE (Beckman Coulter, IM2409), anti-CD34 APC (Beckman Coulter, IM2472U), and anti-CD90 PE (Beckman Coulter, IM1840U). Unstained cells were used as a negative control. For data analysis, the Kaluza 2.0 software (Beckman Coulter) was used.
Cardiogenic differentiation
The differentiation of CMCs toward cardiogenic lineage was done as described.22 The cells were seeded into 60 mm gelatin-coated petri dishes at a density of 10,000 cells/cm2. The next day, the medium was changed to differentiation medium (47% Iscove’s Modified Dulbecco’s Medium, 47% Ham’s F12, 2% horse serum, 1× nonessential amino acids, 1× insulin-transferrin selenium, 1× gentamicin (Invitrogen)) containing 5-azacitidine (10 µM). The next 2 days, 5-azacitidine was added directly to the medium. On the third day, the medium was changed. From the sixth day on, cells were stimulated by ascorbic acid (10−4 M, once every 2 days) and transforming growth factor-β (Peprotech) (1 ng/mL, twice a week). The medium was changed every 2–3 days. Differentiation was carried out for 24 days.
Verification of cardiac-like phenotype after differentiation was done by immunocytochemical (ICC) staining of the cells with anti-α-Actinin Ab (Santa Cruz). Samples were analyzed using a fluorescent microscope, Axio Observer (Carl Zeiss, Germany). Image processing was carried out in the program Axiovision Rel.4.7 (Carl Zeiss, Germany). Cardiac-like phenotype was also verified by quantitative polymerase chain reaction (qPCR) for α-Actinin (ACTN1), NKX2.5, and MEF2C. Examples of cardiogenic-like phenotypes derived from different CMC lines are represented in Supplementary Figs. 3 and 4.
Proliferation assay
Cell proliferation was monitored in real time using the xCELLigence RTCA S16 (Agilent). We seeded 5000 cells in 100 µL of cell suspension in each well of the E-Plate. Then 100 μL of medium was added per well. The impedance value of each well was automatically monitored by the xCELLigence system for a duration of 72 h and expressed as a CI (cell index) value. All samples were analyzed in duplicates. For data analysis, the RTCA Software (version number 1.0.0.1304) was used.
Hypoxic treatment
The cells were placed in a Heracell™ 150 tri-gas incubator (Thermo Scientific, Waltham, MA) set to either 1.0% O2, 5.0% CO2, and 94% N2 or to 5.0% O2, 5.0% CO2, and 90% N2 for the indicated periods (6–72 h). Control CMC conditions included an environment composed of 21% O2, 5.0% CO2, and 74% N2. Cells were then immediately processed for analysis to avoid prolonged exposure to normoxic conditions.
Genetic construct and lentiviruses
Lentiviral packaging plasmids were a generous gift of D. Trono (École Polytechnique Fédérale de Lausanne, Switzerland); pLVTHM was modified to bear Notch1 intracellular domain of Notch1.15 Lentiviral particles were produced using 293 packaging line as described.23 The efficiency of transgene expression with Notch intracellular domain (NICD)-bearing virus was verified by immunoblotting with the Abs to Notch1 (SC6014, Santa Cruz) and by qPCR with primers to Notch target gene HEY1 (data not shown).
qPCR analysis
Total RNA was extracted from CMCs and tissue samples using Extract RNA reagent (Eurogen, Russia) according to the manufacturer’s instructions. Total RNA (1 µg) was reverse transcribed with an MMLV RT Kit (Eurogen, Russia). Real-time PCR was performed with 1 µL cDNA and SYBRGreen PCR Mastermix (Eurogen, Russia) in the Light Cycler system using specific forward and reverse primers for target genes. Primer sequences are shown in Supplementary Table 1. Changes in target gene expression levels were calculated as fold differences using the comparative ∆∆CT method. The mRNA levels were normalized to GAPDH mRNA.
Statistics
Principal component analysis (PCA) was performed to determine which continuous variables discriminate between groups of TF and VSD patients. Continuous variables were qPCR gene expression data from 2−ΔΔCT (RQ) estimation. PCA and the analysis of differential gene expression were performed using the Phantasus tool (https://genome.ifmo.ru/phantasus/) with an integrated limma instrument.24
qPCR data on gene expression was analyzed using GraphPad Prism. Values are expressed as median and interquartile range. The correlation between genes was estimated with Spearman’s Rank coefficient (r). Groups were compared using Mann–Whitney non-parametric test. A value of p ≤ 0.05 was considered significant.
Results
Expression of Notch receptors and ligands in cells from TF and VSD
We obtained cell cultures from myocardial tissues of TF patients and from myocardial tissues of VSD patients used as a comparison group. After three passages in culture, the cells were phenotyped by flow cytometry and demonstrated mesenchymal phenotype (data not shown).
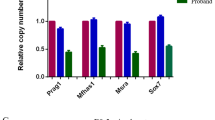
We measured the expression of key genes of the Notch pathway NOTCH1–4, Notch ligands JAG1, DLL4, and Notch target genes HES1, HEY1, HEY2, TWIST, and SNAIL (Fig. 1a). As Bmp2 and Notch cooperate in cardiac patterning,25 we also measured the expression of BMP2. NOTCH4 and DLL4 expression was decreased in the cells of TF compared to VSD cells, whereas NOTCH2, NOTCH3, and JAG1 expression was increased, as well as the expression of target Notch genes HES1, HEY1, and HEY2. The expression of BMP2 was also increased in the cells of TF. Our data suggest alterations of baseline Notch signaling in CMCs of TF patients.
a mRNA levels were analyzed by qPCR and normalized to GAPDH. Vertical axis represents relative mRNA content. Groups are compared using Mann–Whitney non-parametric test; line represents the median; p < 0.05 is considered significant and highlighted by red squares. VSD: n = 14; TF: n = 42. A dot represents mean from qPCR done in duplicates for each individual line of CMC. b Principal component analysis showing differences between CMCs from TF and from VSD by gene expression profiles. c Experimental outline (see “Methods”).
CMCs from TF patients form a cluster by Notch-related gene expression
To identify whether the TF CMCs and VSD CMCs are separated by Notch-related gene expression, we employed PCA and determined which continuous variables discriminate between the groups of VSD and TF. Continuous variables were qPCR gene expression data from 2−ΔΔCT (RQ estimation). Notch-related genes used in PCA were the following: NOTCH1, NOTCH2, NOTCH3, NOTCH4, DLL4, JAG1, HEY2, HES1, HEY1, SNAIL, TWIST1, and BMP2. PCA analysis shows that TF CMCs are indeed separated by gene expression profiles of Notch-related genes from the cells derived from VSD patients (Fig. 1b).
To verify that the found differences were not due to differences in gene expression of CMCs derived from different heart chambers, we isolated CMCs from RAs and RVs of several TF patients and analyzed gene expression by qPCR and conducted PCA analysis as well (Supplementary Fig. 1). We have not found such significant differences between RA- and RV-derived CMCs, especially not in main Notch transcriptional targets. However, we have not observed differences in the expression of Notch-related genes between the tissue samples of RA and RV (Supplementary Fig. 2), which indicates complex cellular content of myocardial tissue.
TF CMCs demonstrated high variability in Notch target gene expression by PCR and elevated NOTCH2,3, and HES1, HEY1, HEY2, as well as BMP2. Therefore, to estimate the overall activation level of Notch signaling in TF CMCs we assessed correlations between NOTCH1, NOTCH2, NOTCH3, and Notch target genes (Fig. 2). We found correlations between the levels of NOTCH1 and targets HES1, HEY1, HEY2; the level of NOTCH3 correlated with HES1. This suggests that Notch1 and Notch3 most likely contribute to the elevated level of Notch signaling seen by HES1, HEY1, and HEY2 in TF CMCs. We observed a moderate correlation between HEY1 and BMP2, raising the question about the cooperation between Bmp2 and Notch in defining the phenotype of TF CMCs. We have not found any associations of high/low Notch signaling with any clinical parameters nor with the age of the patients (data not shown).
The level of Notch activation is associated with proliferation in TF CMCs
We observed high variability in Notch activation level among TF CMCs (Fig. 1). We hypothesized that TF CMCs could have altered stemness potency not only depending on age but also depending on the level of Notch activation. To verify this, we first analyzed whether age or high/low Notch level could be associated with a higher/lower proliferation rate of CMCs from TF patients. We divided CMCs from TF patients by their age: neonates <14 days, 5–7 months, and >12 months. This age division was made because of several factors. We analyzed proliferation of corresponding CMC lines using xCELLigence real-time cell analysis assay26 (Fig. 3a) and did not observe any association of age group with proliferation activity of CMCs. In contrast, when we divided CMC lines by their Notch activity by HEY1/HES1 expression, we observed that higher Notch signaling was associated with a higher rate of proliferation, and lower Notch activity was associated with a lower proliferation rate, correspondingly (Fig. 3b).
Cell proliferation was assessed by the xCELLigence real-time cell analysis assay; n = 16. a CMC proliferation and age of TF patients. Dotted plot represents age distribution of TF patients; n = 42. CMCs representing three age periods are indicated by different colors. Corresponding growth curves are represented on the right plot. b CMC proliferation and Notch activation of TF patients. Dotted plots represent the expression levels of HES1 and HEY1 by qPCR in the CMC of TF patients. CMCs representing “low level” and “high level” are indicated by blue and red, respectively. Corresponding growth curves are represented on the right plot. Numbers with arrows show patient IDs. Horizontal lines indicate median values, n = 24.
The level of Notch activation is associated with higher differentiation potency of TF CMCs
The functional outcome of Notch signaling activation is highly dependent on the developmental context,27 and conflicting data exist on the activating and inactivating roles of Notch in various types of differentiation, including differentiation of cardiac lineages.28,29 Therefore, we analyzed whether various levels of Notch signaling influenced the differentiation potency of CMCs from TF. Cardiogenic-like capacities have been previously described for CMCs.22 We verified cardiogenic properties of CMC lines with ICC staining and with qPCR for cardiogenic markers (Supplementary Figs. 3 and 4)
We induced cardiogenic-like differentiation in the lines of TF CMCs and divided into four groups with various levels of Notch activation by NOTСH1/HEY1 (Fig. 4). The level of BMP2 expression corresponded to those of HEY1/NOTCH1 (Fig. 4a). Twenty-four days after the induction of cardiogenic differentiation, we observed an increase in the expression levels of cardiogenic markers MEF2C and NKX2-5 to the extent corresponding to the initial level of Notch activation (Fig. 4b). The results from this and the previous section suggest that an elevated level of Notch signaling could contribute to the increased plasticity of CMCs of TF patients.
a Four CMC lines from TF patients were chosen by their different levels of Notch activation. Corresponding levels of NOTCH1, HEY1, and BMP2 by qPCR are represented. b Cardiogenic differentiation was induced in the four CMC lines. The levels of MEF2C and NKX2-5 were assessed by qPCR 24 days after the induction of differentiation. The experiments were done in triplicate; error bars reflect mean +/− SD of triplicates.
Notch signaling dose-dependently promotes differentiation
To confirm that elevated levels of Notch signaling could contribute to elevated potency of CMCs to differentiate to cardiogenic-like lineage, we activated Notch signaling in CMC via lentiviral transduction with viruses bearing activating NICD15 and induced cardiogenic differentiation, respectively. Transduction with NICD-bearing virus induced dose-dependent elevation of NOTCH1 and HEY1 expression in CMCs (Fig. 5a). NICD induced elevation in the level of cardiogenic markers MEF2C and NKX2-5 in a nice dose-dependent manner (Fig. 5b), suggesting the specific action of NICD. In NICD-transduced CMCs, more intensive ICC staining for cardiaс marker α-Actinin was observed after 24 days of cardiogenic differentiation (Fig. 5c). The response of CMC lines from independent TF patients to NICD activation is shown in Supplementary Fig. 5. Thus Notch activation increases the differentiation potency of CMCs.
a NICD-bearing viruses were added to CMCs (1 and 10 µL, correspondingly; C control). Expression of NOTCH1 and HEY1 was verified by qPCR. b Cardiogenic differentiation was induced in five individual CMC lines transduced with different amounts of NICD-bearing viruses. Histograms represent MEF2C and NKX2-5 expression by qPCR at day 24 after the induction of cardiogenic differentiation. Error bars reflect mean +/− SD of triplicates. *,**p < 0.05 in relation to control samples. c Immunocytochemical staining of the CMC for a cardiogenic marker α-Actinin in the presence/absence of Notch activation by NICD (NICD and C, respectively) in undifferentiated and differentiated cells. The experiments were done in triplicate; individual biological replicates for CMCs from different patients are represented in Supplementary Figs. 3 and 4.
Hypoxic injury moderately elevates Notch signaling
TF is the most common cyanotic CHD. This pathology includes a wide spectrum of severity: from pink TF without hypoxemia and almost normal arterial saturation up to very cyanotic neonates with a very low level of saturation. More than half of TF patients have a reduced level of saturation (hypoxemia). As a result, all tissues and organs of the body suffer from hypoxia, including brain and myocardial tissue. The role of Notch signaling in myocardial remodeling in hypoxic conditions is not clear. We hypothesized that the hypoxic condition could itself be a cause of elevated Notch signaling in the cells of TF patients. To verify this, we cultured CMCs from a VSD patient in hypoxic conditions using 1% and 5% O2, correspondingly, and used culture in normoxic condition (21% O2) as a control. Culture of CMCs in hypoxic conditions caused just a very moderate elevation of NOTCH1 and HEY1 transcription as well as the transcription of BMP2 (Fig. 6a). These data suggest that hypoxic conditions in TF patients could potentially contribute to an activation of Notch signaling in CMCs.
a CMCs were cultured in hypoxic conditions using 1% (1% hypoxia) and 5% (5% hypoxia) O2, correspondingly, and normoxic condition (21% O2) as a control, C. Histograms represent NOTCH1, HEY1, and BMP2 expression by qPCR. Groups are compared using Mann–Whitney non-parametric test; five individual lines of CMCs from VSD were used in the experiment; the experiment was repeated three times; *p < 0.05. b Analysis of Notch-activated genes in CMCs. NICD-bearing viruses (1 and 10 µL, correspondingly; C control) were added to CMCs or to HUVECs used as a non-cardiogenic control. Histograms represent corresponding gene expression 48 h after the viral transduction by qPCR. Three individual lines of CMCs from VSD were used in the experiment. The experiments were done in triplicate. Individual biological replicates for CMCs from different patients are represented in Supplementary Fig. 5.
Concerted but independent action of Notch and bone morphogenetic protein 2 (BMP2) has been reported for mouse heart development.25 To verify whether the expression of BMP2 along with cardiogenic markers was dependent on Notch activation, we activated Notch in CMCs by NICD-bearing lentiviruses and observed dose-dependent elevation of NOTCH1, HEY1, MEF2C, and NKX2.5. HUVECs were used as a non-cardiogenic control. We did not observe elevation in BMP2 transcription by NICD in CMCs (Fig. 6b), suggesting that the mechanism of BMP2 activation in CMCs is not dependent on Notch activation.
Discussion
TF is the most common, severe cyanotic congenital heart defect. Cellular and molecular mechanisms of this developmental defect remain unknown. In this study, we show a change in the expression pattern of Notch-related genes in CMCs of patients with TF compared to cells derived from VSD, known not to be related to Notch signaling when it is an isolated defect. Notch-related genes demonstrated high variability of the level of expression in TF cells from low to high expression. We have not found any associations of high/low Notch signaling with any clinical parameters nor with the age of patients. A high level of Notch activation was associated with higher proliferation of cells and also with their higher potency to differentiation.
No single gene locus, with the exception of the 22q11 deletion, has been found to be associated with TF cases.4 A recent study, which is the largest whole-exome sequencing investigation of sporadic, non-syndromic TF performed to date, showed that the NOTCH1 locus is the most frequent site of genetic variants predisposing to non-syndromic TF. A total of 829 non-syndromic TF patients underwent whole-exome sequencing for 77 previously described genes. NOTCH1 was most frequently found to harbor deleterious variants. Variants were not confined to a single functional domain of the NOTCH1 protein, but significant clustering of variants was evident in the epidermal growth factor (EGF)-like repeats.10 EGF-like repeats of Notch receptors are required for ligand binding and external signalizations between the cells.30 Other most frequent variants identified in TF patients are related to vascular endothelial growth factor signaling.10 These findings indicate that pathogenic mechanisms of TF are related to signaling taking part in vessel formation and endothelial regulation.
Apart from genetic studies, some attempts have been made to analyze a resulting outcome in the heart tissues of TF patients. In a study that included both idiopathic TF patients and patients with syndromic 22q11.2 deletion, >1000 dysregulated genes were identified in the TF-derived RV tissues of both patient groups.9 The expression of the majority of genes associated with the Wnt and Notch signaling pathway were significantly reduced. The comparison of idiopathic and syndromic patients suggests that different genetic starting conditions may nevertheless lead to a common pathological outcome.
Heart development is a complex morphogenetic process that depends on the precise spatial and temporal coordination of many signaling pathways. Notch often acts in concert with other signaling pathways, especially Wnt and BMP, to regulate these processes. Recent data on the role of Notch signaling in cardiac development illustrate how the Notch pathway functions as a signal integrator of tissue patterning during outflow tract development by regulating cell proliferation, differentiation, and apoptosis.31
It has been shown that global and tissue-specific NOTCH1 heterozygous mice could be phenotypically normal, with no obvious cardiovascular pathologies.32 However, in a more recent study that assessed ascending aortic aneurysm, NOTCH1+/− in a predominantly 129S6 background developed aortic root dilation; this was in contrast to NOTCH1+/− in a mixed background.33 These findings highlight the importance of genetic background in disease expressivity, are consistent with the incomplete penetrance observed, and also are well in line with our observations of the broad variability of Notch-related gene expression in the cells of TF patients. However, in our study, we cannot exclude other factors relating to specific genotypes of the TF patients or technical noise introduced during CMC establishment.
Hypoxic conditions have been described as a Notch-modulating factor.34 TF patients suffer from hypoxia; however, our data show only a slight increase in Notch-related gene expression and in the expression of BMP2 in response to induced hypoxia. We suggest that this could indicate that the changes in Notch-related gene expression and BMP2 expression observed in the cells of TF patients are rather related to a changed genetic background of these patients than to hypoxic conditions.
It has been shown recently that Bmp2 is able to activate the Notch pathway during early murine heart development.25 Ectopic Bmp2 expression in the mouse chamber myocardium was able to change the transcriptional signature of adjacent chamber endocardial cells into valve tissue and enabled them to undergo epithelial-to-mesenchymal transition,25 a process critical for the whole-heart patterning and proper chamber development.7 We suggest that the altered expression level of Notch components and BMP2 observed in the CMCs of TF patients could reflect pathogenic events related to impaired cardiac patterning in TF patients.
Cardiac progenitor/mesenchymal cells have been used recently in several clinical trials to improve univentricular heart function mostly in patients with hypoplastic left heart syndrome35 and are in general considered a promising source of cardiac multipotent cells. The trials gave some promising results and initiated research on stemness properties of CMC and related cells.19,20 These latter studies highlight the importance of the patient’s age when considering stemness of CMCs. However, the obvious decline in stemness properties of CMCs has been shown for patients of age <1 month to >40 years in a recent study,19 whereas another paper shows that the cardiomyogenic potential of cardiosphere-derived cells is uniform during the first year of life.20
TF patients are mostly of a very young age at surgical intervention, so we had only a restricted opportunity to analyze the effect of age on proliferation of CMCs and have not revealed any influence of either the age or any clinical parameters of TF patients on CMC proliferation. However, our data suggest that the higher the level of Notch pathway activity, the greater the proliferation and differentiation capacity of CMCs from the corresponding TF patients. Our observations are well in line with studies demonstrating involvement of Notch activation in proliferation and differentiation properties of stem cells.29,36,37
Notably, tissue samples demonstrated a different expression profile when compared to CMC gene expression (Supplementary Figs. 1 and 2). On one hand, this is a limitation in extrapolating data from tissue samples to cell subsets building the mature heart. But on the other hand, this is the first study in which CMC and heart tissue samples from the RAs and RVs of the same patients were directly compared. We suggest that our study contributes to a deeper understanding of CMC biology.
Thus we suggest that our observations on cellular effects of low/high Notch activation on CMC plasticity might be important for future development of cellular therapy strategies.
Surgical correction of TF has been well established for the past approximately 40 years, and as a result, survival has improved dramatically: 90% of patients are currently alive 30 years after successful surgical correction at a young age.38 Despite these satisfactory results, survival up to 30 years is lower than in the normal population. Although anatomic correction and physiological correction have been achieved, complications such as pulmonary regurgitation leading to RV dysfunction, recurrent obstruction of the RV outflow tract, arrhythmias, sudden death, and aortic dilation and regurgitation are found in late survivors.39 This raises the important question of whether impaired genetic background, including Notch gene dysregulation, still influences TF patients even after surgical correction.
Our data on expression profiles in TF patients reveal Notch dysregulation rather than overall downregulation. This dysregulation could be responsible for the early events of the heart patterning common for all TF events.
We recently reported dysregulation of Notch genes in the cells of patients with thoracic aortic aneurysm, bicuspid aortic valve, and aortic valve calcification regardless of the presence of mutations in the NOTCH1 gene.15,16,17,40 All together, our observations of Notch dysregulation in several cardiac pathologies raise the possibility that dysregulated Notch gene expression is associated with a pathological state of a broad spectrum of heart tissues.41 Complex genetic background could converge on Notch signaling as a very sensitive and fine-tuned regulating component of tissue integrity. As a result, dysregulation in the expression of Notch-related genes causes failure of a complex and balanced regulating mechanism, and this in total could lead to developmental defects not necessarily associated with mutations in Notch-related genes.
Our study highlights the importance of Notch activation level in CMCs of patients with TF. However, an important limitation of the study is the lack of data on cells derived from normal myocardium in young patients. Owing to obvious reasons, it is difficult to obtain any age-matched young control of similar myocardial tissue biopsies or biopsies from young children a few years after corrective surgery to verify our findings. Another limitation concerns lack of direct sequencing data on NOTCH1 and does not exclude single variants in our population. However, major syndromic forms were excluded from the study. Another limitation is the fact that we were unable to have CMC and tissue samples from all patients and also from both RA and RV. Clearly, further research is needed for a deeper understanding of the role of various factors in TF pathogenesis as well as the factors influencing CMC biology.
References
Hoffman, J. I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002).
Lahm, H. et al. Tetralogy of Fallot and hypoplastic left heart syndrome–complex clinical phenotypes meet complex genetic networks. Curr. Genomics 16, 141–158 (2015).
Bailliard, F. & Anderson, R. H. Tetralogy of Fallot. Orphanet J. Rare Dis. 4, 2 (2009).
Mercer-Rosa, L. et al. 22q11. 2 deletion status and disease burden in children and adolescents with tetralogy of Fallot. Circ. Cardiovasc. Genet. 8, 74–81 (2015).
Töpf, A. et al. Functionally significant, rare transcription factor variants in tetralogy of Fallot. PLoS ONE 9, e95453 (2014).
Morgenthau, A. & Frishman, W. H. Genetic origins of tetralogy of Fallot. Cardiol. Rev. 26, 86–92 (2018).
de la Pompa, J. L. & Epstein, J. A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22, 244–254 (2012).
Greenway, S. C. et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat. Genet. 41, 931 (2009).
Bittel, D. C. et al. Gene expression in cardiac tissues from infants with idiopathic conotruncal defects. BMC Med. Genomics 4, 1 (2011).
Page, D. J. et al. Whole exome sequencing reveals the major genetic contributors to nonsyndromic tetralogy of Fallot. Circ. Res. 124, 553–563 (2019).
Andersson, E. R., Sandberg, R. & Lendahl, U. Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612 (2011).
Luxán, G., D’Amato, G., MacGrogan, D. & de la Pompa, J. L. Endocardial Notch signaling in cardiac development and disease. Circ. Res. 118, e1–e18 (2016).
Maleki, S. et al. Mesenchymal state of intimal cells may explain higher propensity to ascending aortic aneurysm in bicuspid aortic valves. Sci. Rep. 6, 35712 (2016).
Balistreri, C. R. et al. Deregulation of Notch1 pathway and circulating endothelial progenitor cell (EPC) number in patients with bicuspid aortic valve with and without ascending aorta aneurysm. Sci. Rep. 8, 13834 (2018).
Kostina, A. S. et al. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim. Biophys. Acta 1862, 733–740 (2016).
Kostina, A. et al. Different Notch signaling in cells from calcified bicuspid and tricuspid aortic valves. J. Mol. Cell Cardiol. 114, 211–219 (2018).
Kostina, A. et al. Notch, BMP and WNT/beta-catenin network is impaired in endothelial cells of the patients with thoracic aortic aneurysm. Atheroscler. Suppl. 35, e6–e13 (2018).
Menasche, P. Cell therapy trials for heart regeneration - lessons learned and future directions. Nat. Rev. Cardiol. 15, 659–671 (2018).
Sharma, S. et al. A deep proteome analysis identifies the complete secretome as the functional unit of human cardiac progenitor cells. Circ. Res. 120, 816–834 (2017).
Traister, A. et al. Cardiac regenerative capacity is age- and disease-dependent in childhood heart disease. PLoS ONE 13, e0200342 (2018).
Zaidi, S. & Brueckner, M. Genetics and GENOMICS OF CONGENITAL HEART DISease. Circ. Res. 120, 923–940 (2017).
Smits, A. M. et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat. Protoc. 4, 232–243 (2009).
Malashicheva, A., Kanzler, B., Tolkunova, E., Trono, D. & Tomilin, A. Lentivirus as a tool for lineage‐specific gene manipulations. Genesis 45, 456–459 (2007).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Papoutsi, T., Luna-Zurita, L., Prados, B., Zaffran, S. & de la Pompa, J. L. Bmp2 and Notch cooperate to pattern the embryonic endocardium. Development 145, dev163378 (2018).
Dowling, C. M., Herranz Ors, C. & Kiely, P. A. Using real-time impedance-based assays to monitor the effects of fibroblast-derived media on the adhesion, proliferation, migration and invasion of colon cancer cells. Biosci. Rep. 34, e00126 (2014).
Bray, S. J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 (2016).
Boni, A. et al. Notch1 regulates the fate of cardiac progenitor cells. Proc. Natl Acad. Sci. USA 105, 15529–15534 (2008).
Gude, N. et al. Notch activation enhances lineage commitment and protective signaling in cardiac progenitor cells. Basic Res. Cardiol. 110, 1–15. (2015).
Kovall, R. A., Gebelein, B., Sprinzak, D. & Kopan, R. The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell 41, 228–241 (2017).
MacGrogan, D., Munch, J. & de la Pompa, J. L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 15, 685–704 (2018).
Conlon, R. A., Reaume, A. G. & Rossant, J. Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545 (1995).
Koenig, S. N. et al. Notch1 haploinsufficiency causes ascending aortic aneurysms in mice. JCI Insight 2, 91353 (2017).
Borggrefe, T. et al. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim. Biophys. Acta 1863, 303–313 (2016).
Sano, T. et al. Impact of cardiac progenitor cells on heart failure and survival in single ventricle congenital heart disease. Circ. Res. 117, 312311 (2018).
Urbanek, K. et al. Inhibition of Notch1-dependent cardiomyogenesis leads to a dilated myopathy in the neonatal heart. Circ. Res. 107, 429–441 (2010).
Collesi, C. et al. Reversible Notch1 acetylation tunes proliferative signalling in cardiomyocytes. Cardiovasc. Res. 114, 103–122 (2018).
Geva, T. Indications and timing of pulmonary valve replacement after tetralogy of Fallot repair. Semin. Thorac. Cardiovasc. Surg. Pediatr. Card. Surg. Annu. 11–22 (2006).
Dluzniewska, N. et al. Long-term follow-up in adults after tetralogy of Fallot repair. Cardiovasc. Ultrasound 16, 28 (2018).
Ignatieva, E. et al. Mechanisms of smooth muscle cell differentiation are distinctly altered in thoracic aortic aneurysms associated with bicuspid or tricuspid aortic valves. Front. Physiol. 8, 536 (2017).
Aquila, G. et al. The Notch pathway: a novel therapeutic target for cardiovascular diseases? Expert Opin. Ther. Targets 23, 695–710 (2019).
Acknowledgements
This study was supported by a grant from Russian Science Foundation 19-74-00092.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.M., I.K.; data curation: A. Kostina, A. Kiselev; formal analysis: E.I., A.G.; funding acquisition: A.M., A. Kostareva; investigation: I.K., P.D., A.M.; methodology: E.I.; project administration: A.M.; resources: T.P., E.G., M.G.; software: A. Kostina, A. Kiselev; supervision: A.M., A. Kostareva; writing—original draft; A.M., I.K.; writing—review and editing: A.M., I.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kozyrev, I., Dokshin, P., Kostina, A. et al. Dysregulation of Notch signaling in cardiac mesenchymal cells of patients with tetralogy of Fallot. Pediatr Res 88, 38–47 (2020). https://doi.org/10.1038/s41390-020-0760-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0760-6