Abstract
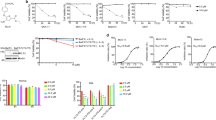
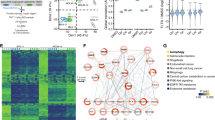
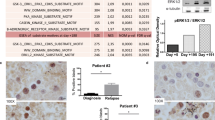
The internal tandem duplication of the FMS-like tyrosine kinase 3 (FLT3-ITD) is one of the most frequent genetic alterations in acute myeloid leukemia (AML). Limited and transient clinical benefit of FLT3 kinase inhibitors (FLT3i) emphasizes the need for alternative therapeutic options for this subset of myeloid malignancies. Herein, we showed that FLT3-ITD mutant (FLT3-ITD+) AML cells were susceptible toward inhibitors of DHODH, a rate-limiting enzyme of de novo pyrimidine biosynthesis. Genetic and pharmacological blockade of DHODH triggered downregulation of FLT3-ITD protein, subsequently suppressed activation of downstream ERK and STAT5, and promoted cell death of FLT3-ITD+ AML cells. Mechanistically, DHODH blockade triggered autophagy-mediated FLT3-ITD degradation via inactivating mTOR, a potent autophagy repressor. Notably, blockade of DHODH synergized with an FDA-approved FLT3i quizartinib in significantly impairing the growth of FLT3-ITD+ AML cells and improving tumor-bearing mice survival. We further demonstrated that DHODH blockade exhibited profound anti-proliferation effect on quizartinib-resistant cells in vitro and in vivo. In summary, this study demonstrates that the induction of degradation of FLT3-ITD protein by DHODH blockade may offer a promising therapeutic strategy for AML patients harboring FLT3-ITD mutation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published paper and its supplementary information file.
References
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21.
Nitika, Wei J, Hui AM. Role of Biomarkers in FLT3 AML. Cancers. 2022;14:1164.
Newell LF, Cook RJ. Advances in acute myeloid leukemia. BMJ (Clin Res ed). 2021;375:n2026.
Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312.
Kazi JU, Rönnstrand L. FMS-like Tyrosine Kinase 3/FLT3: From Basic Science to Clinical Implications. Physiol Rev. 2019;99:1433–66.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10.
Long J, Jia MY, Fang WY, Chen XJ, Mu LL, Wang ZY, et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood. 2020;135:1472–83.
Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–31.
Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med. 2017;377:454–64.
Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. N Engl J Med. 2019;381:1728–40.
Cortes JE, Khaled S, Martinelli G, Perl AE, Ganguly S, Russell N, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20:984–97.
Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood. 2009;114:2984–92.
McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov. 2019;9:1050–63.
Alotaibi AS, Yilmaz M, Kanagal-Shamanna R, Loghavi S, Kadia TM, DiNardo CD, et al. Patterns of Resistance Differ in Patients with Acute Myeloid Leukemia Treated with Type I versus Type II FLT3 inhibitors. Blood Cancer Discov. 2021;2:125–34.
Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–3.
Williams AB, Nguyen B, Li L, Brown P, Levis M, Leahy D, et al. Mutations of FLT3/ITD confer resistance to multiple tyrosine kinase inhibitors. Leukemia. 2013;27:48–55.
Weisberg E, Ray A, Nelson E, Adamia S, Barrett R, Sattler M, et al. Reversible resistance induced by FLT3 inhibition: a novel resistance mechanism in mutant FLT3-expressing cells. PloS ONE. 2011;6:e25351.
Albers C, Leischner H, Verbeek M, Yu C, Illert AL, Peschel C, et al. The secondary FLT3-ITD F691L mutation induces resistance to AC220 in FLT3-ITD+ AML but retains in vitro sensitivity to PKC412 and Sunitinib. Leukemia. 2013;27:1416–8.
Reiter K, Polzer H, Krupka C, Maiser A, Vick B, Rothenberg-Thurley M, et al. Tyrosine kinase inhibition increases the cell surface localization of FLT3-ITD and enhances FLT3-directed immunotherapy of acute myeloid leukemia. Leukemia. 2018;32:313–22.
Sugamori H, Lee T, Mitomi T, Yamagishi C. Interim results from a postmarketing surveillance study of patients with FLT3-mutated relapsed/refractory AML treated with the FLT3 inhibitor gilteritinib in Japan. Jpn J Clin Oncol. 2022;52:766–73.
Kiyoi H. FLT3 INHIBITORS: Recent Advances And Problems For Clinical Application. Nagoya J Med Sci. 2015;77:7–17.
Azhar M, Kincaid Z, Kesarwani M, Ahmed A, Wunderlich M, Latif T, et al. Momelotinib is a highly potent inhibitor of FLT3-mutant AML. Blood Adv. 2022;6:1186–92.
Kohli L, Kaza N, Coric T, Byer SJ, Brossier NM, Klocke BJ, et al. 4-Hydroxytamoxifen induces autophagic death through K-Ras degradation. Cancer Res. 2013;73:4395–405.
Wu H, Lu XX, Wang JR, Yang TY, Li XM, He XS, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15:1506–22.
Löffler M, Jöckel J, Schuster G, Becker C. Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol Cell Biochem. 1997;174:125–9.
Christian S, Merz C, Evans L, Gradl S, Seidel H, Friberg A, et al. The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia. 2019;33:2403–15.
Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell. 2016;167:171–86.e15.
So J, Lewis AC, Smith LK, Stanley K, Franich R, Yoannidis D, et al. Inhibition of pyrimidine biosynthesis targets protein translation in acute myeloid leukemia. EMBO Mol Med. 2022;14:e15203.
Peters GJ, Schwartsmann G, Nadal JC, Laurensse EJ, van Groeningen CJ, van der Vijgh WJ, et al. In vivo inhibition of the pyrimidine de novo enzyme dihydroorotic acid dehydrogenase by brequinar sodium (DUP-785; NSC 368390) in mice and patients. Cancer Res. 1990;50:4644–9.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16.
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84.
Breedveld FC, Dayer JM. Leflunomide: mode of action in the treatment of rheumatoid arthritis. Ann Rheum Dis. 2000;59:841–9.
Cao L, Weetall M, Trotta C, Cintron K, Ma J, Kim MJ, et al. Targeting of Hematologic Malignancies with PTC299, A Novel Potent Inhibitor of Dihydroorotate Dehydrogenase with Favorable Pharmaceutical Properties. Mol Cancer Ther. 2019;18:3–16.
McDonald G, Chubukov V, Coco J, Truskowski K, Narayanaswamy R, Choe S, et al. Selective Vulnerability to Pyrimidine Starvation in Hematologic Malignancies Revealed by AG-636, a Novel Clinical-Stage Inhibitor of Dihydroorotate Dehydrogenase. Mol Cancer Ther. 2020;19:2502–15.
Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin-proteasome system: collaborators in neuroprotection. Biochim et Biophys Acta. 2008;1782:691–9.
Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502.
Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12.
Sudarsanam S, Johnson DE. Functional consequences of mTOR inhibition. Curr Opin Drug Discov Dev 2010;13:31–40.
Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41.
Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402.
Jenö P, Ballou LM, Novak-Hofer I, Thomas G. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA. 1988;85:406–10.
Brunn GJ, Hudson CC, Sekulić A, Williams JM, Hosoi H, Houghton PJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101.
Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–7.
Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94.
Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34.
Jetani H, Garcia-Cadenas I, Nerreter T, Goetz R, Sierra J, Bonig H, et al. FLT3 Inhibitor Treatment Increases FLT3 Expression That Exposes FLT3-ITD+ AML Blasts to Elimination By FLT3 CAR-T Cells. Blood. 2018;132:903.
Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinforma (Oxf, Engl). 2016;32:2866–8.
Ianevski A, Giri AK, Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50:W739–w43.
Weisberg EL, Schauer NJ, Yang J, Lamberto I, Doherty L, Bhatt S, et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat Chem Biol. 2017;13:1207–15.
Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19:55–60.
Green AS, Maciel TT, Hospital MA, Yin C, Mazed F, Townsend EC, et al. Pim kinases modulate resistance to FLT3 tyrosine kinase inhibitors in FLT3-ITD acute myeloid leukemia. Sci Adv. 2015;1:e1500221.
Fröhling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–80.
Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–9.
Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–42.
Döhner H, Wei AH, Löwenberg B. Towards precision medicine for AML. Nat Rev Clin Oncol. 2021;18:577–90.
Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem. 2004;279:33035–8.
Shi DD, Savani MR, Levitt MM, Wang AC, Endress JE, Bird CE, et al. De novo pyrimidine synthesis is a targetable vulnerability in IDH mutant glioma. Cancer Cell. 2022;40:939–56.e16.
Li L, Ng SR, Colón CI, Drapkin BJ, Hsu PP, Li Z, et al. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci Transl Med. 2019;11:eaaw7852.
Wu D, Wang W, Chen W, Lian F, Lang L, Huang Y, et al. Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica. 2018;103:1472–83.
Zhou J, Yiying Quah J, Ng Y, Chooi JY, Hui-Min Toh S, Lin B, et al. ASLAN003, a potent dihydroorotate dehydrogenase inhibitor for differentiation of acute myeloid leukemia. Haematologica. 2020;105:2286–97.
Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab. 2017;25:1037–43.
Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42.
Kang C, Avery L. To be or not to be, the level of autophagy is the question: dual roles of autophagy in the survival response to starvation. Autophagy. 2008;4:82–4.
Mao J, Ma L, Shen Y, Zhu K, Zhang R, Xi W, et al. Arsenic circumvents the gefitinib resistance by binding to P62 and mediating autophagic degradation of EGFR in non-small cell lung cancer. Cell Death Dis 2018;9:963.
Garufi A, Pucci D, D’Orazi V, Cirone M, Bossi G, Avantaggiati ML, et al. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis 2014;5:e1271.
Booth L, Roberts JL, Poklepovic A, Kirkwood J, Sander C, Avogadri-Connors F, et al. The levels of mutant K-RAS and mutant N-RAS are rapidly reduced in a Beclin1 / ATG5 -dependent fashion by the irreversible ERBB1/2/4 inhibitor neratinib. Cancer Biol Ther. 2018;19:132–7.
Schlessinger J. Signal transduction. Autoinhibition control. Science. 2003;300:750–2.
Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–78.
Larrue C, Saland E, Boutzen H, Vergez F, David M, Joffre C, et al. Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood. 2016;127:882–92.
Moore AS, Faisal A, Gonzalez de Castro D, Bavetsias V, Sun C, Atrash B, et al. Selective FLT3 inhibition of FLT3-ITD+ acute myeloid leukaemia resulting in secondary D835Y mutation: a model for emerging clinical resistance patterns. Leukemia. 2012;26:1462–70.
He X, Zhu Y, Lin YC, Li M, Du J, Dong H, et al. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD(+) acute myeloid leukemia. Blood. 2019;134:548–60.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (81973362, 82173857), Shanghai Committee of Science and Technology (22S11900900), Guangdong Provincial Medical Science and Technology Research Fund (B2021077) and Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism. We thank the Ministry of Education and the Research Center of Analysis and Test of East China University of Science and Technology.
Author information
Authors and Affiliations
Contributions
This work was conceptualized by HM, JYC, ZHL, WQL, and JH. Mouse model were performed by HM, JYC, ZHL. Cell culture, qPCR, confocal analysis, were done by HM, ZHL. Immunoblot analysis, electron microscopy analysis and plasmid construction experiments were performed by HM, ZHL, WQF, SSL, SYC, and YYZ. BAY-234 was provided by JAC, LXL, YHW, and QX. Bioinformatics analysis were done by HM and ZHL. Data analysis and interpretation were done by HM, JYC, ZHL, KFL, and HYT. The manuscript was drafted by HM, JYC, WQL, and JH, and reviewed and modified by all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal studies and protocols were approved by the Animal Experiments Committee of East China University of Science and Technology (ECUST-2021-03007).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, H., Cui, J., Liu, Z. et al. Blockade of de novo pyrimidine biosynthesis triggers autophagic degradation of oncoprotein FLT3-ITD in acute myeloid leukemia. Oncogene 42, 3331–3343 (2023). https://doi.org/10.1038/s41388-023-02848-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02848-7