Abstract
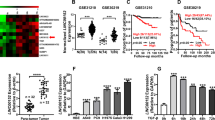
Metastasis is responsible for most cancer mortality, but its molecular mechanism has not been completely understood. In addition to coding genes and miRNAs, the contribution of long noncoding RNAs (lncRNAs) to tumor metastatic dissemination and the mechanisms controlling their expression are areas of intensive investigation. Here, we show that lncRNA NORAD is downregulated in lung and breast cancers, and that NORAD low expression in these cancer types is associated with lymph node metastasis and poor prognosis. NORAD is transcriptionally repressed by the Hippo pathway transducer YAP/TAZ-TEAD complex in conjunction with the action of NuRD complex. Functionally, NORAD elicits potent inhibitory effects on migration and invasion of multiple lung and breast cancer cell lines, and repression of NORAD expression participates in the migration- and invasion-stimulatory effects of the YAP pathway. Mechanistically, NORAD exploits its multiple repeated sequences to function as a multivalent platform for binding and sequestering S100P, thereby suppressing S100P-elicited pro-metastatic signaling network. Using cell and mouse models, we show that the S100P decoy function of NORAD suppresses lung and breast cancer migration, invasion, and metastasis. Together, our study identifies NORAD as a novel metastasis suppressor, elucidates its regulatory and functional mechanisms, and highlights its prognostic value.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–40.
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208.
Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19.
Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–38.
Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–51.
Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712.
de Andres-Pablo A, Morillon A, Wery M. LncRNAs, lost in translation or licence to regulate? Curr Genet. 2017;63:29–33.
Sun M, Kraus WL. From discovery to function: the expanding roles of long non-coding RNAs inphysiology and disease. Endocr Rev. 2015;36:25–64.
Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209.
Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80.
Tichon A, Perry RB, Stojic L, Ulitsky I. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev. 2018;32:70–8.
Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561:132–6.
Prica F, Radon T, Cheng Y, Crnogorac-Jurcevic T. The life and works of S100P - from conception to cancer. Am J Cancer Res. 2016;6:562–76.
Arumugam T, Logsdon CD. S100P: a novel therapeutic target for cancer. Amino Acids. 2011;41:893–9.
Whiteman HJ, Weeks ME, Dowen SE, Barry S, Timms JF, Lemoine NR, et al. The role of S100P in the invasion of pancreatic cancer cells is mediated through cytoskeletal changes and regulation of cathepsin D. Cancer Res. 2007;67:8633–42.
Heil A, Nazmi AR, Koltzscher M, Poeter M, Austermann J, Assard N, et al. S100P is a novel interaction partner and regulator of IQGAP1. J Biol Chem. 2011;286:7227–38.
Du M, Wang G, Ismail TM, Gross S, Fernig DG, Barraclough R, et al. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem. 2012;287:15330–44.
Austermann J, Nazmi AR, Muller-Tidow C, Gerke V. Characterization of the Ca2+ -regulated ezrin-S100P interaction and its role in tumor cell migration. J Biol Chem. 2008;283:29331–40.
Gibadulinova A, Pastorek M, Filipcik P, Radvak P, Csaderova L, Vojtesek B, et al. Cancer-associated S100P protein binds and inactivatesp53, permits therapy-induced senescence and supports chemoresistance. Oncotarget. 2016;7:22508–22.
Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803.
Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–312.
Valencia-Sama I, Zhao Y, Lai D, Janse van Rensburg HJ, Hao Y, Yang X. Hippo component TAZ functions as a co-repressor and negatively regulates DeltaNp63 transcription through TEA Domain (TEAD) transcription factor. J Biol Chem. 2015;290:16906–17.
Kim M, Kim T, Johnson RL, Lim DS. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11:270–82.
Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. The TEAD family and its oncogenic role in promoting tumorigenesis. Int J Mol Sci. 2016;17:E138.
Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74.
Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5.
Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–80.
Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix MJ, Wu R, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17:353–60.
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61.
Das A, Fischer RS, Pan D, Waterman CM. YAP nuclear localization in the absence of cell-cell contact is mediated by a filamentous actin-dependent, myosin II- and phospho-YAP-independent pathway during extracellular matrix mechanosensing. J Biol Chem. 2016;291:6096–110.
Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–51.
Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–14.
Kim JK, Jung KH, Noh JH, Eun JW, Bae HJ, Xie HJ, et al. Targeted disruption of S100P suppresses tumor cell growth by down-regulation of cyclin D1 and CDK2 in human hepatocellular carcinoma. Int J Oncol. 2009;35:1257–64.
Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol. 2017;92:34–42.
Wu X, Lim ZF, Li Z, Gu L, Ma W, Zhou Q, et al. NORAD expression is associated with adverse prognosis in esophageal squamous cell carcinoma. Oncol Res Treat. 2017;40:370–4.
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169.
Kawasaki N, Miwa T, Hokari S, Sakurai T, Ohmori K, Miyauchi K, et al. Long noncoding RNA NORAD regulates transforming growth factor-beta signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci. 2018;109:2211–20.
Elster D, Tollot M, Schlegelmilch K, Ori A, Rosenwald A, Sahai E, et al. TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells. Nat Commun. 2018;9:3115.
Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J, et al. The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell. 2016;167:1525–39 e1517.
von Eyss B, Jaenicke LA, Kortlever RM, Royla N, Wiese KE, Letschert S, et al. A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell. 2015;28:743–57.
Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S, et al. A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun. 2015;6:10186.
Zhang Z, Du J, Wang S, Shao L, Jin K, Li F, et al. OTUB2 promotes cancer metastasis via Hippo-independent activation of YAP and TAZ. Mol Cell. 2019;73:7–21 e27.
Wu HC, Lin YC, Liu CH, Chung HC, Wang YT, Lin YW, et al. USP11 regulates PML stability to control Notch-induced malignancy in brain tumours. Nat Commun. 2014;5:3214.
Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, et al. A Cullin3-KLHL20 ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20:214–28.
Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, et al. K33-linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell. 2014;54:586–600.
Acknowledgments
We thank Sherry Hsueh-Chi Yen and Pan-Chyr Yang for reagents, Suh-Yuen Liang for TCGA analysis, National RNAi Core Facility for shRNA constructs and CRISPR vectors and constructs, Academia Sinica Animal Core Facility for IVIS analysis, Institute of Biological Chemistry Histopathology Core Facility for tissue processing and histology, Academia Sinica Common Mass Spectrometry Facilities for MS analysis, Human Biobank, Research Center of Clinical Medicine, and Cancer Data Bank of National Cheng Kung University Hospital for patient’s specimens. This work is supported by National Health Research Institute Grant NHRI-EX107-10708BI and intramural fund from Institute of Biological Chemistry, Academia Sinica.
Author information
Authors and Affiliations
Contributions
RHC conceived the project. BST, MCY, and RHC designed experiments. BST, MCY, SS, HYC, and YCC conducted experiments. MYW and YCW provided breast and lung cancer patient specimens, respectively.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tan, BS., Yang, MC., Singh, S. et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38, 5612–5626 (2019). https://doi.org/10.1038/s41388-019-0812-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0812-8
This article is cited by
-
Role of ferroptosis and ferroptosis-related long non'coding RNA in breast cancer
Cellular & Molecular Biology Letters (2024)
-
Metformin and long non-coding RNAs in breast cancer
Journal of Translational Medicine (2023)
-
The interplay between noncoding RNA and YAP/TAZ signaling in cancers: molecular functions and mechanisms
Journal of Experimental & Clinical Cancer Research (2022)
-
The biology of YAP in programmed cell death
Biomarker Research (2022)
-
Long noncoding RNA BCRP3 stimulates VPS34 and autophagy activities to promote protein homeostasis and cell survival
Journal of Biomedical Science (2022)