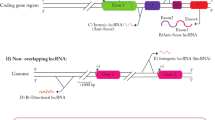
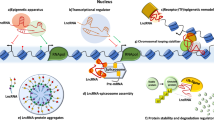
Abstract
Metastasis is a major contributor to cancer-associated deaths. It is characterized by a multistep process that occurs through the acquisition of molecular and phenotypic changes enabling cancer cells from a primary tumour to disseminate and colonize at distant organ sites. Over the past decade, the discovery and characterization of long noncoding RNAs (lncRNAs) have revealed the diversity of their regulatory roles, including key contributions throughout the metastatic cascade. Here, we review how lncRNAs promote metastasis by functioning in discrete pro-metastatic steps including the epithelial–mesenchymal transition, invasion and migration and organotrophic colonization, and by influencing the metastatic tumour microenvironment, often by interacting within ribonucleoprotein complexes or directly with other nucleic acid entities. We discuss well-characterized lncRNAs with in vivo phenotypes and highlight mechanistic commonalities such as convergence with the TGFβ–ZEB1/ZEB2 axis or the nuclear factor-κB pathway, in addition to lncRNAs with controversial mechanisms and the influence of methodologies on mechanistic interpretation. Furthermore, some lncRNAs can help identify tumours with increased metastatic risk and spur novel therapeutic strategies, with several lncRNAs having shown potential as novel targets for antisense oligonucleotide therapy in animal models. In addition to well-characterized examples of lncRNAs functioning in metastasis, we discuss controversies and ongoing challenges in lncRNA biology. Finally, we present areas for future study for this rapidly evolving field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Amin, M. B. AJCC Cancer Staging Manual. 8th Edn. (Springer, 2017).
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis. Cell 168, 670–691 (2017).
Valastyan, S. & Weinberg, R. A. Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 (2011). Seminal primer on invasion–metastasis cascade and pathogenesis of tumour metastasis.
Gupta, G. P. & Massagué, J. Cancer metastasis: building a framework. Cell 127, 679–695 (2006).
Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013). Comprehensive review of lncRNA concepts.
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009).
St Laurent, G., Wahlestedt, C. & Kapranov, P. The landscape of long noncoding RNA classification. Trends Genet. 31, 239–251 (2015).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011). Establishes the widespread and cell type-specific nature of lncRNA expression.
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015).
FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Engreitz, J. M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016).
Barr, M. L. & Bertram, E. G. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163, 676–677 (1949).
Chen, C.-K. et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472 (2016).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Willingham, A. T. et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573 (2005).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Lee, S. et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). Establishes HOTAIR as an important epigenetic regulator in breast cancer metastasis.
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Tsai, M.-C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Bond, A. M. et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027 (2009).
Berghoff, E. G. et al. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140, 4407–4416 (2013).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014).
Trimarchi, T. et al. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606 (2014).
Xiang, J.-F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014).
Ma, W. et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 12, 71–78 (2015).
Latos, P. A. et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012).
Kornienko, A. E., Guenzl, P. M., Barlow, D. P. & Pauler, F. M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11, 59 (2013).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Mele, M. & Rinn, J. L. ‘Cat’s cradling’ the 3D genome by the act of LncRNA transcription. Mol. Cell 62, 657–664 (2016).
Yao, R.-W., Wang, Y. & Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551 (2019).
Kim, J. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 (2018). Challenges the concept of MALAT1 as a pro-metastasis lncRNA by revealing an alternative molecular mechanism.
Cho, S. W. et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22 (2018).
Keniry, A. et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14, 659–665 (2012).
Schmitt, A. M. & Chang, H. Y. Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016).
Slack, F. J. & Chinnaiyan, A. M. The role of non-coding RNAs in oncology. Cell 179, 1033–1055 (2019).
Hu, X. et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357 (2014).
Tseng, Y.-Y. et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014). Establishes genomic alterations of lncRNA loci as important features in cancer.
McCleland, M. L. et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Invest. 126, 639–652 (2016).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Yin, Y. et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516 (2015).
Paralkar, V. R. et al. Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110 (2016).
Groff, A. F. et al. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 16, 2178–2186 (2016).
Dimitrova, N. et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790 (2014).
Huarte, M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010).
Weidle, U. H., Birzele, F., Kollmorgen, G. & Rüger, R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteom. 14, 143–160 (2017).
Huang, Q., Yan, J. & Agami, R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev. 37, 75–81 (2018).
Li, J., Meng, H., Bai, Y. & Wang, K. Regulation of lncRNA and its role in cancer metastasis. Oncol. Res. 23, 205–217 (2016).
Raab, J. R. et al. SWI/SNF remains localized to chromatin in the presence of SCHLAP1. Nat. Genet. 51, 26–29 (2019).
Liu, S. J. & Lim, D. A. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 19, e46955 (2018).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111 (2017).
Liu, Y. et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat. Biotechnol. 1656, 175–1210 (2018).
Zhu, S. et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat. Biotechnol. 34, 1279–1286 (2016).
Boettcher, M. et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat. Biotechnol. 36, 170–178 (2018).
Liu, S. J. et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 21, 83 (2020).
Thiery, J. P., Acloque, H., Huang, R. Y. J. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Nieto, M. A., Huang, R. Y. J., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Yang, J. et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
De Craene, B. & Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 13, 97–110 (2013).
Brannan, C. I., Dees, E. C. & Ingram, R. S. The product of the H19 gene may function as an RNA. Mol. Cell. 10, 28–36 (1990).
Rainier, S. et al. Relaxation of imprinted genes in human cancer. Nature 362, 747–749 (1993). One of the earliest reports of a noncoding RNA involved in cancer.
Luo, M. et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333, 213–221 (2013).
Davidovich, C., Zheng, L., Goodrich, K. J. & Cech, T. R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257 (2013).
Liang, W.-C. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525 (2015).
Zhou, W. et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 10, eaak9557 (2017). Demonstrates the role of H19 in a stepwise cascade of metastasis initiation and colonization, with alternative molecular mechanisms based on the cellular context.
Zhu, M. et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281, 3766–3775 (2014).
Chen, W.-Y. et al. Loss of SPDEF and gain of TGFBI activity after androgen deprivation therapy promote EMT and bone metastasis of prostate cancer. Sci. Signal 10, eaam6826 (2017).
Liang, H. et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer 17, 119–13 (2018).
Grelet, S. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19, 1105–1115 (2017). A well dissected example of a lncRNA acting as a ceRNA to regulate the EMT.
Yuan, J.-H. et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014).
Shi, S.-J. et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 6, 11652–11663 (2015).
Wu, N. et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 65, 87–14 (2020).
Li, T. et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 75, 3181–3191 (2015).
Liu, F. et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 35, 5422–5434 (2016).
Wang, H. et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 16, 136 (2017).
Li, Y. et al. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J. Cell. Biochem. 120, 10633–10642 (2019).
Terashima, M., Tange, S., Ishimura, A. & Suzuki, T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem. 292, 82–99 (2017).
Mondal, T. et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6, 7743–17 (2015).
Yan, X. et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res. 77, 6704–6716 (2017).
Mitra, R. et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun. 8, 1604–1612 (2017).
Li, Z. et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 24, 59–71 (2017).
Li, W. et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Invest. 127, 3421–3440 (2017).
Liang, W.-C. et al. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene 37, 1445–1456 (2018).
Tichon, A. et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 7, 12209 (2016).
Tan, B.-S. et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38, 5612–5626 (2019).
Ling, H. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013).
Redis, R. S. et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol. Cell 61, 520–534 (2016).
Silva-Fisher, J. M. et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat. Commun. 11, 2156–13 (2020). A lncRNA that is both prognostic of survival in metastatic colon cancer and predictive of response to chemotherapy and targeted molecular therapy.
Ji, P. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Arun, G. et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016).
Gutschner, T. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 (2013). Seminal loss of function study of MALAT1 as a lncRNA regulator of cancer metastasis.
Zhang, B. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012).
Sterling, J. A. et al. The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression and osteolysis in metastatic human breast cancer cells. Cancer Res. 66, 7548–7553 (2006).
Xing, Z. et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125 (2014).
Meijer, D., van Agthoven, T., Bosma, P. T., Nooter, K. & Dorssers, L. C. J. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol. Cancer Res. 4, 379–386 (2006).
Calon, A. et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012).
Kogo, R. et al. Long noncoding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011).
Ren, Y. et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ß1 secretion. Mol. Cancer 17, 5–14 (2018).
Gupta, G. P. et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb. Symposia Quant. Biol. 70, 149–158 (2005).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Yuzhalin, A. E. & Yu, D. Brain metastasis organotropism. Cold Spring Harb. Perspect. Med. 10, a037242 (2020).
Weidle, U. H., Birzele, F., Kollmorgen, G. & Rüger, R. Molecular basis of lung tropism of metastasis. Cancer Genomics Proteom. 13, 129–139 (2016).
Shimo, T. et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J. Bone Miner. Res. 21, 1045–1059 (2006).
Chen, W., Hoffmann, A. D., Liu, H. & Liu, X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2, 4 (2018).
Guy, C. T., Cardiff, R. D. & Muller, W. J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992).
Muller, W. J., Sinn, E., Pattengale, P. K., Wallace, R. & Leder, P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105–115 (1988).
Annunziato, S., Barazas, M., Rottenberg, S. & Jonkers, J. Genetic dissection of cancer development, therapy response, and resistance in mouse models of breast cancer. Cold Spring Harb. Symposia Quant. Biol. 81, 141–150 (2016).
Li, C. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 19, 106–119 (2017). Studies the role of MAYA in directing organ-specific tropism towards bone metastases and demonstrates therapeutic in vivo targeting of this lncRNA in mice.
Valiente, M. et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014).
Wang, S. et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Invest. 127, 4498–4515 (2017). Demonstrates how lnc-BM promotes brain metastases through activation of cell adhesion molecules and cytokine release.
Lee, J. T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12, 815–826 (2011).
Xing, F. et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78, 4316–4330 (2018).
De Palma, M., Biziato, D. & Petrova, T. V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017).
Kitamura, T., Qian, B.-Z. & Pollard, J. W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 15, 73–86 (2015).
Karki, R. & Kanneganti, T.-D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 19, 197–214 (2019).
Huntington, N. D., Cursons, J. & Rautela, J. The cancer-natural killer cell immunity cycle. Nat. Rev. Cancer 7, 703–718 (2020).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP and TAZ: a signalling hub of the tumour microenvironment. Nat. Rev. Cancer 19, 454–464 (2019).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Liu, B. et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381 (2015).
Huang, D. et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol. 19, 1112–1125 (2018). Broadens the role of NKILA toinclude both tumour-intrinsic and tumour microenvironment factors.
Ji, J. et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 9, 478 (2018).
Sang, L.-J. et al. LncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol. Cell 72, 71–83.e7 (2018).
Jiang, R. et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 8, 15129 (2017).
Chen, C. et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 9, 3826–18 (2018).
Chen, Y. G., Satpathy, A. T. & Chang, H. Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972 (2017).
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2014).
Sauvageau, M. et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2, e01749 (2013). Genetic deletion of multiple lncRNAs in mouse models demonstrating essential phenotypes.
Roobol, M. J. et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur. Urol. 58, 475–481 (2010).
Prensner, J. R. et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 (2014). Large-scale translational study establishing SChLAP1 as a prognostic marker for metastatic prostate cancer risk.
Prensner, J. R. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398 (2013).
Reyes, D. K. & Pienta, K. J. The biology and treatment of oligometastatic cancer. Oncotarget 6, 8491–8524 (2015).
Palma, D. A. et al. The oligometastatic state - separating truth from wishful thinking. Nat. Publ. Group. 11, 549–557 (2014).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016). Demonstrates paracrine transfer of lncRNAs through exosomes leading to drug resistance.
Yoshida, T. et al. Estimated number of off-target candidate sites for antisense oligonucleotides in human mRNA sequences. Genes Cell 23, 448–455 (2018).
Yoshida, T. et al. Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cell 24, 827–835 (2019).
Lee, J.-S. & Mendell, J. T. Antisense-mediated transcript knockdown triggers premature transcription termination. Mol. Cell 77, 1044–1054.e3 (2020).
Lai, F., Damle, S. S., Ling, K. K. & Rigo, F. Directed RNase H cleavage of nascent transcripts causes transcription termination. Mol. Cell 77, 1032–1043.e4 (2020).
Swayze, E. E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007).
Meng, L. et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412 (2015).
Scharner, J. et al. Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res. 48, 802–816 (2020).
Zielinski, R. & Chi, K. N. Custirsen (OGX-011): a second-generation antisense inhibitor of clusterin in development for the treatment of prostate cancer. Future Oncol. 8, 1239–1251 (2012).
Yamamoto, Y. et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin. Cancer Res. 21, 1675–1687 (2015).
Finkel, R. S. et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017). Randomized clinical trial of an antisense oligonucleotide-based therapeutic in human patients demonstrating successful clinical response.
Hong, D. et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 7, 314ra185 (2015).
Monteleone, G. et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N. Engl. J. Med. 372, 1104–1113 (2015).
Cornelis, G., Souquere, S., Vernochet, C., Heidmann, T. & Pierron, G. Functional conservation of the lncRNA NEAT1 in the ancestrally diverged marsupial lineage: evidence for NEAT1 expression and associated paraspeckle assembly during late gestation in the opossum Monodelphis domestica. RNA Biol. 13, 826–836 (2016).
Karner, H. et al. Functional conservation of LncRNA JPX despite sequence and structural divergence. J. Mol. Biol. 432, 283–300 (2020).
Long, Y. et al. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 52, 931–938 (2020).
Fares, J., Fares, M. Y., Khachfe, H. H., Salhab, H. A. & Fares, Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal. Transduct. Target. Ther. 5, 28–17 (2020).
Bassett, A. R. et al. Considerations when investigating lncRNA function in vivo. eLife 3, e03058 (2014).
Wurster, C. D. & Ludolph, A. C. Antisense oligonucleotides in neurological disorders. Ther. Adv. Neurol. Disord. 11, 1756286418776932 (2018).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283 (2016).
Denzler, R., Agarwal, V., Stefano, J., Bartel, D. P. & Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 (2014).
Ala, U. et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl Acad. Sci. USA 110, 7154–7159 (2013).
Figliuzzi, M., Marinari, E. & De Martino, A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys. J. 104, 1203–1213 (2013).
Hausser, J. & Zavolan, M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat. Rev. Genet. 15, 599–612 (2014).
Thomson, D. W., Bracken, C. P. & Goodall, G. J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 39, 6845–6853 (2011).
Gennarino, V. A. et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 22, 1163–1172 (2012).
Du, Z. et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 7, 10982–10 (2016).
Paraskevopoulou, M. D. et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 41, D239–D245 (2013).
Wang, P. et al. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database 2015, bav098 (2015).
Hoffmann, M. et al. SPONGEdb: a pan-cancer resource for competing endogenous RNA interactions. NAR Cancer https://doi.org/10.1093/narcan/zcaa042 (2021).
Li, J.-H., Liu, S., Zhou, H., Qu, L.-H. & Yang, J.-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 (2014).
Karreth, F. A. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 (2015).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cancer thanks M. Huarte, L. Ma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Invasion–metastasis cascade
-
A stepwise succession of events whereby primary tumour cells adopt a phenotype that promotes local invasion, intravasation into the bloodstream, extravasation and migration to distant organ sites, and colonization of those sites, which often involves physiological adaptation.
- Long noncoding RNAs
-
(lncRNAs). Noncoding RNA transcripts longer than 200 nucleotides, which are frequently polyadenylated, and that show no evidence that they encode proteins.
- Competing endogenous RNAs
-
(ceRNAs). RNA transcripts (which may be lncRNAs, mRNAs or pseudogenes) that are capable of binding and influencing the activity of microRNAs through complementary base pairing.
- Organ-specific tropism
-
The propensity for disseminated cancer cells to colonize and proliferate at specific organ sites due to diverse physiological and molecular factors.
- Antisense oligonucleotides
-
(ASOs). Exogenous oligonucleotides, often chemically modified to resist degradation, that alter the amount, stability or activity of complementary RNA transcripts, usually through RNase-based mechanisms.
- Epithelial–mesenchymal transition
-
(EMT). A set of molecular processes that convert cancer cells of polarized epithelial phenotypes into more invasive mesenchymal phenotypes.
- Anoikis
-
A subtype of programmed cell death triggered by inappropriate cellular or extracellular matrix interactions, often in the setting of tumour cells escaping their primary environment.
- Micrometastases
-
Collections of metastasized tumour cells that are clinically detectable but typically under 2 mm in diameter in human patients.
- Comprehensive identification of RNA-binding proteins by mass spectrometry
-
(ChIRP-MS). A method of identifying endogenous RNA–protein interactions with high specificity, using in vivo crosslinking, RNA pulldown and mass spectrometry.
Rights and permissions
About this article
Cite this article
Liu, S.J., Dang, H.X., Lim, D.A. et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer 21, 446–460 (2021). https://doi.org/10.1038/s41568-021-00353-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-021-00353-1
This article is cited by
-
The role of ADAR1 through and beyond its editing activity in cancer
Cell Communication and Signaling (2024)
-
Cancer-derived exosomes as novel biomarkers in metastatic gastrointestinal cancer
Molecular Cancer (2024)
-
Ascites exosomal lncRNA PLADE enhances platinum sensitivity by inducing R-loops in ovarian cancer
Oncogene (2024)
-
LncRNA PRBC induces autophagy to promote breast cancer progression through modulating PABPC1-mediated mRNA stabilization
Oncogene (2024)
-
FAK-LINC01089 negative regulatory loop controls chemoresistance and progression of small cell lung cancer
Oncogene (2024)