Abstract
ADHD is a highly prevalent neurodevelopmental disorder. The first-line therapeutic for ADHD, methylphenidate, can cause serious side effects including weight loss, insomnia, and hypertension. Therefore, the development of non-stimulant-based therapeutics has been prioritized. However, many of these also cause other effects, most notably somnolence. Here, we have used a uniquely powerful genetic model and unbiased drug screen to identify novel ADHD non-stimulant therapeutics. We first found that adgrl3.1 null (adgrl3.1−/−) zebrafish larvae showed a robust hyperactive phenotype. Although the hyperactivity was rescued by three ADHD non-stimulant therapeutics, all interfered significantly with sleep. Second, we used wild-type zebrafish larvae to characterize a simple behavioral phenotype generated by atomoxetine and screened the 1200 compound Prestwick Chemical Library® for a matching behavioral profile resulting in 67 hits. These hits were re-assayed in the adgrl3.1−/−. Using the previously identified non-stimulants as a positive control, we identified four compounds that matched the effect of atomoxetine: aceclofenac, amlodipine, doxazosin, and moxonidine. We additionally demonstrated cognitive effects of moxonidine in mice using a T-maze spontaneous alternation task. Moxonidine, has high affinity for imidazoline 1 receptors. We, therefore, assayed a pure imidazoline 1 agonist, LNP599, which generated an effect closely matching other non-stimulant ADHD therapeutics suggesting a role for this receptor system in ADHD. In summary, we introduce a genetic model of ADHD in zebrafish and identify five putative therapeutics. The findings offer a novel tool for understanding the neural circuits of ADHD, suggest a novel mechanism for its etiology, and identify novel therapeutics.
Similar content being viewed by others
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is a neurodevelopmental disorder with a prevalence of approximately 2–5% worldwide [1]. Symptoms appear as early as in preschool, with some patients showing pervasive behavioral and psychiatric deficits into adulthood [2]. ADHD is often associated with functional and psychosocial comorbidities as well as a higher risk of developing mood and anxiety disorders [3]. When left unmanaged, the consequences of ADHD range from impaired quality of life to risk of suicide [4].
While stimulants, including methylphenidate and d-amphetamines, are considered first-line therapeutics for ADHD [5,6,7,8,9], they are associated with adverse reactions such as decreased appetite, headaches, and sleep disturbances [10] and have high abuse liability [11]. This has fueled the development of non-stimulants as ADHD therapeutics [12]. Non-stimulants, such as atomoxetine, guanfacine, clonidine, and viloxazine [13] are considered second-line ADHD therapeutics. Non-stimulants are also commonly associated with adverse events such as fatigue, somnolence, agitation, and aggression and they tend to be less effective than stimulants [14]. Importantly, about one-quarter of patients do not benefit from treatment with neither stimulants nor non-stimulants [7, 15]. Thus, there is an urgent need to develop novel therapeutics for ADHD.
Family, sibling, and adoption studies suggest strong heritability for ADHD [16, 17] and hundreds of relevant genetic markers have been revealed using candidate gene association, genome-wide association, and copy number variant studies [18]. Of the genes identified, LPHN3 has been most robustly linked to ADHD [19,20,21,22]. The ADGRL3 gene codes for an adhesion G protein-coupled receptor; it has an important role in plasticity, cell adhesion, and synapse formation and is expressed in key brain regions involved with attention [23, 24]. Variants in ADGRL3 are statistically associated with increased risk of ADHD [25]. This association was replicated in a case-control association study [26]. The ADGRL3 risk haplotype significantly affects neural function [27] and has been shown to cause both hyperactivity and cognitive deficits [24, 28] in rats and mice [29]. Similarly, studies in larval zebrafish have demonstrated that morpholino oligonucleotide knockdown of adgrl3.1 function leads to a hyperactive and impulsive motor phenotype [30]. The motor phenotype observed in adgrl3.1 morphants was rescued by treatment with methylphenidate and atomoxetine [30]. Biological validation of mutations in ADRGL3 from humans to rodent and zebrafish demonstrates the usefulness of these translational models in ADHD drug discovery [31,32,33].
The aim of this study was to screen FDA-approved compounds for therapeutic effects in ADHD. To that end, we introduce a novel CRISPR-cas9 adgrl3.1 mutant model of ADHD in zebrafish and benchmarked the ensuing drug screen against the non-stimulant atomoxetine [30]. First, a simple behavioral phenotype generated by atomoxetine was characterized in wild-type larvae. Next, we screened 1200 compounds for a matching phenotype and re-assayed the hits in the adgrl3.1 mutant model. Five novel ADHD candidate therapeutics were identified and one was also shown to match the effects of atomoxetine in a cognitive mouse assay.
Methods and materials
Fish
For behavioral analysis, a total of N = 12480 6 days-post-fertilization (dpf) mutant and N = 56528 wild-type larvae were used (all group sizes are reported in supplementary tables 1–6). Wildtype zebrafish (AB line) were initially obtained from ZIRC (Eugene, OR, USA). All zebrafish were maintained in the laboratory at the University of Reykjavik. Zebrafish were fed three times a day on a variable diet of TetraMin flakes (Tetra Holding GmbH, Melle, Germany), Adult Zebrafish Complete Diet (Zeigler Bros, Gardners, PA, USA), and live Artemia (INVE Aquaculture, Incorporation, Salt Lake City, UT, USA). Fish were kept in a 14:10 light:dark cycle (lights-on at 8:00 am) in 10 L multi-tank constant flow system tanks (Aquatic Habitats, Apopka, FL, USA). Water temperature was held at a constant of 28.5 °C and replaced at a rate of 10% per day. All procedures in this study were carried out in strict compliance with the regulations of and approved by, the National Bioethics Committee of Iceland (regulation 460/2017).
Generation of adgrl3.1 zebrafish knock out using CRISPR/Cas9
The adgrl3.1 mutant line (adgrl3.1−/− are referred to as homozygous mutants and adgrl3.1+/+ as WT) was generated as described in [34]. A short guide RNA (sgRNA) was designed containing a PAM motif targeting all the splice variants of adgrl3.1 shown in the ENSEMBL database: Adgrl3.1-201 exon 11 to 13; Adgrl3.1-202 exon 10 to 12; Adgrl3.1-203 exon 5 to 7; Adgrl3.1-204 exon 9 to 11; Adgrl3.1-205 exon 11 to 13; Adgrl3.1-206 exon 11 to 13. The sgRNA sequences that we used were sgRNA-1: GAGTCCTCCAGTCTGATAGG and sgRNA-2: GCAAGAAGTGTGGGTGCGGT.
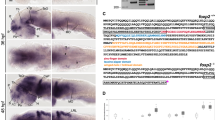
The gRNA constructs were co-injected at a concentration of 100 ng/µl together with 50 ng/nl Cas9 protein (Fig. 1A–C).
A Cartoon showing position of forward and reverse sgRNA constructs used to create the novel adgrl3.1−/− line. B The base pair sequence of adgrl3.1 that is deleted by sgRNA injection. The nucleotides shown in orange were not removed and so are retained in the mutant version of adgrl3.1. C The CRISPR guide RNAs were designed to target the different splice variants of adgrl3.1 found in the Ensembl database. (NMD: non-sense mediated decay). D Swim velocity of homozygous wild type (WT, n = 184) and homozygous adgrl3.1−/− (HOM, n = 179) larvae throughout the whole recording period. The data are pooled from four experiments. The time between 13:30 and 18:00 shows alternating intervals of 30-min lights-off (grey shaded bar) and lights-on phases, followed by constant light-on from 18:00 to 22:00. For the night time, from 22:00 to 08:00 the next morning, the lights were turned off (grey shaded bar). E The average distance moved for the five 30-min lights-off periods demonstrates that homozygous adgrl3.1−/− larvae moved significantly more than wild-type larvae. F Sleep parameters, expressed as fold change of wild-type larvae, were examined throughout the night, during 10-h lights-off phase. No significant differences between genotypes were observed in any of the five sleep parameters. * denote significant differences.
We screened for founder fish by in-crossing putative founders and collecting the eggs. Genomic DNA was extracted from these eggs by phenol-chloroform extraction and re-suspended in distilled water. We used the following primers to amplify a fragment of the adgrl3.1 gene: Forward TGTGCTTTGGTGTGGGTGCTAATGTA and Reverse GGATGCAAAACAGGGTGGGTTGAGGG. 1 µl of 30 ng/µl genomic DNA was used as template in a PCR reaction together with 18.9 µl dH2O, 4 µl Phusion HF buffer, 0.4 µl 10 mM dNTP mix, 0.5 µl primer mix (30 ng/µl each) and 0.2 µl Phusion polymerase. The PCR program used the following parameters: 98 °C 3 min; 40 cycles of [98 °C 30 s; 68 °C 30 s; 72 °C 1.30 min]; 72 °C 5 min and 7 °C hold. The completed PCR reactions were loaded onto a 1% agarose gel containing 5 µl Tris-Borate-EDTA (TBE) in a 100 ml gel. Mutations were identified by a shift in the size of the band produced by PCR on the agarose gel. Promising founders were grown up to adulthood and the nature of the deletion identified by sequencing. The novel adgrl3.1 mutant line was generated by selecting a founder fish containing a 650 bp deletion that leads frameshift mutations in each splice variant of adgrl3.1.
Genotyping
adgrl3.1+/- carriers were identified by fin clipping potential carriers and extracting DNA by either phenol-chloroform extraction or using the HotShot method. A fragment of the adgrl3.1 gene was amplified using the protocol above, and mutations identified by either gel electrophoresis or Sanger sequencing.
Behavioral Recordings
Behavioral assays were performed using a custom-built activity monitoring system, as previously described [35, 36]. At 5 dpf, larvae were individually placed in 96-microwell plates (Nunc, Roskilde, Denmark) in system water and acclimated in the system for 24 h. All recordings started at 13:00 with the lights on and continued with alternating dark and light phases in 30-min intervals between 13:30 and 18:00 for the adgrl3.1 (−/− and +/+) strain but with uninterrupted 14:10 light-dark cycles for wild-types. The behavior was tracked at 5 Hz and larvae that tracked <90% of the total recording time were excluded; each larvae received only one drug/dose
Motor Assay
Three behavioral parameters were calculated: Average velocity during day and night. Peak velocity (mm/s), defined as the average highest velocity of the five light-to-dark transitions, measured for a 30 s period immediately following the change from light to dark. And distance moved (mm), defined as the average distance moved during the five 30-min light phases.
Sleep assay
Sleep behavior [37] was recorded in a 96-well plate and analyzed during the lights-off period (22:00 - 08:00). All procedures have been described previously [36, 38, 39]. Briefly, behavioral states were dichotomized into 1-s bins of movement/non-movement (0.5 cm/s set as the threshold for movement). All activity that was slower than that threshold was described as non-movement. After six or more consecutive 1-s bins of non-movement, the seventh second and above were classified as sleep; all other bouts were classified as wake.
Drug preparation and administration
All compounds were dissolved in dimethyl sulfoxide (DMSO) and subsequently diluted to reach a final concentration between 0.1 and 100 µM in the system water, using 0.3% DMSO as control. Wild-type larvae were assayed using the 1200 compound Prestwick Chemical Library® (Prestwick Chemical, Strasbourg, France) in three concentrations 1, 10, and 35 µM. Mutant larvae were re-assayed using a 67-compound subset of the library in 1, 10, and 30 µM concentrations. Five compounds: aceclofenac, amlodipine, doxazosin, moxonidine (Sigma-Aldrich, St. Louis, MO, USA), and LNP599 (Greenpharma, Orléans, France) were repeated in five concentrations 0.1, 1, 10, 30, and 100 µM. All compounds were prepared the day before recording and administered into the wells between 11:30 and 12:30 on the day of recording.
Mice
Seventy (male only, 30.6 ± 0.2 g) CD-1 mice (Janvier Labs, Le Genest Saint Isle, France) aged 4-5 weeks were used. The study was conducted with the approval of the Institutional Animal Care and Use Committees (CREMEAS (CEEA 35), Strasbourg, France) and in compliance with European legislation on animal care and scientific experimentation (Permit no: 16808-2018092015164989v3). Mice were group-housed in an enriched environment (sizzle dry, tunnel, and smart house) and maintained in a room with controlled temperature (21–22 °C) and a reversed light-dark cycle (12 h/12 h; lights on: 17:30–05:30; lights off: 05:30–17:30) with food and water available ad libitum.
T-maze spontaneous alternation assay
T-maze spontaneous alternation is an established assay to assess cognitive performance in rodents [40]. The alternation performance is drastically reduced with administration of drugs such as scopolamine and this effect is reversible with cognitive-enhancing [41, 42] drugs. The T-maze assay and apparatus has been described previously [43]. Seven groups of mice (n = 10) were used: (1) Vehicle + Saline; (2) Vehicle + Scopolamine; (3) Scopolamine + Donepezil (0.3 mg/kg); (4) Scopolamine + Atomoxetine (3 mg/kg); (5) Scopolamine + Moxonidine (0.1 mg/kg); (6) Scopolamine + Moxonidine (0.3 mg/kg) and (7) Scopolamine + Moxonidine (1 mg/kg). Scopolamine, Atomoxetine, and Donepezil were used at 1, 3, and 0.3 mg/kg, respectively. All compounds were dissolved in saline and were administered with scopolamine using i.p. injection 30 min prior to the assay except for donepezil which was administrated p.o. 60 min prior to assay. (Scopolamine and Atomoxetine: Sigma-Aldrich, Saint-Quentin-Fallavier, France; Donepezil: Tocris Biotechene, Noyal Chatillon sur Seiche, France).
Data analysis
Data was obtained using EthoVision XT (Version 11.5.2016, Noldus) and exported to Microsoft Excel for motility analysis and custom-written software for sleep analysis. Statistical analysis was performed using GraphPad Prism Software (Version 8.4.3, GraphPad Software Inc.). For behavioral analysis of zebrafish, statistical differences were evaluated using an unpaired t-test and a Bonferroni correction between wild-type and homozygous adgrl3.1−/− mutant larvae for all parameters. Statistical differences between compound-treated groups were evaluated using one-way ANOVA and Dunnett’s multiple comparison post hoc analysis. P < 0.05 was considered statistically significant. The percentage of spontaneous alternations was calculated as the number of spontaneous alternations divided by 14 (number of free-choice trials). Data was then analyzed using unpaired t-test to evaluate the difference between vehicle + saline and vehicle-+ scopolamine groups and one-way ANOVA with Dunnett´s multiple comparison post hoc test to evaluate the difference between vehicle + scopolamine and the different compound + scopolamine groups. All data are presented as mean ± standard error (s.e.m).
Results
adgrl3.1 mutants exhibit robust hyperactivity phenotype compared to wild-type larvae
adgrl3.1−/− larvae were consistently hyperactive compared to their wild type controls while their sleep patterns did not differ (Fig. 1D). Both the average velocity during day (0.874 ± 0.035 mm/s vs. 0.690 ± 0.027 mm/s, t(360) = 4.148, p < 0.001), and the peak velocity was higher following an abrupt day-time lights-off stimulus (1.632 ± 0.047 mm/s vs. 1.445 ± 0.045 mm/s, t(360) = 2.883, p < 0.01). However, the velocity during night did not differ (n.s.). The difference in average velocity re-emerged after lights-on (08:00); (0.672 ± 0.032 mm/s vs. 0.535 ± 0.024 mm/s, t(360) = 3.454, p < 0.001)(Fig. 1E). An independent samples t-test revealed a significant difference between genotypes for the average distance moved during the day-time 30-min lights-on bouts, where adgrl3.1 homozygous mutant larvae moved significantly more (1590 ± 38.25 vs. 1204 ± 26.75 mm/30 min, t(361) = 8.298, p < 0.001) than the wild type (Fig. 1F).
Statistical analysis did not reveal a difference between genotypes for any of the five sleep variables (Fig. 1F). We conclude that the average distance moved during daytime 30-min lights-on bouts (hereafter average distance moved; Cohen´s d equals 0.87) is a robust parameter that differentiates well between mutants and wild-types and represents an ideal parameter for drug screening.
Atomoxetine, clonidine, and guanfacine rescue the motility phenotype of adgrl3.1 mutants and interfere with sleep parameters
To assess the validity of the hyperactive adgrl3.1−/− zebrafish model, we assessed the effects of three non-stimulant ADHD therapeutics on the average distance moved and sleep (Fig. 2A–D). One-way ANOVA revealed a statistically significant difference between adgrl3.1−/− control (DMSO) group and adgrl3.1−/− larvae treated with clonidine (F(3, 186) = 26.46, p < 0.001), atomoxetine (F(3, 258) = 76.36, p < 0.001) and guanfacine (F(3, 187) = 63.65, p < 0.001) for the average distance moved. Dunnett´s post hoc test revealed that all three drugs, lowered the average distance moved for all concentrations tested, 1 µM, 10 µM, and 30 µM, compared to the control group (Supplementary Table 1). In brief, all ADHD therapeutics tested rescued the motility phenotype of the mutant larvae.
adgrl3.1−/− larvae were treated with three different concentrations (1 µM, 10 µM, and 30 µM) of A clonidine, B atomoxetine, and C guanfacine and compared to larvae treated with 0.3% DMSO. Average distance moved during the five 30-min periods of lights-on and sleep parameters during the night was analyzed. Clonidine, atomoxetine and guanfacine all reduced the distance moved during lights-on significantly for all three concentrations. Clonidine, atomoxetine, and guanfacine all significantly altered the sleep dynamics at multiple parameters during the night. Wild-type (WT) larvae treated with 0.3% DMSO are included for visual comparison. * denote significant differences.
All three drugs affected sleep parameters of compound-treated larvae compared to adgrl3.1−/− control group (Fig. 2A–C). One-way ANOVA revealed that clonidine had a significant effect on all five sleep parameters, sleep fragmentation (F(3,185) = 14.342, p < 0.001), sleep ratio (F(3,185) = 32.65, p < 0.001), velocity (F(3,185) = 84.397, p < 0.001), wake bout duration (F(3,185) = 40.562, p < 0.001) and sleep bout duration (F(3,185) = 24.443, p < 0.001). Atomoxetine: sleep fragmentation (F(3,255) = 25.208, p < 0.001), sleep ratio (F(3,255) = 32.237, p < 0.001), velocity (F(3,255) = 17.464, p < 0.001), wake bout duration (F(3, 255) = 4.829, p < 0.001) and sleep bout duration (F(3, 255) = 26.296, p < 0.001). Guanfacine: (F(3,185) = 25.531, p < 0.001), velocity (F(3,185) = 37.473, p < 0.001), wake bout duration (F(3,185) = 31.125, p < 0.001) and sleep bout duration (F(3,185) = 16.337, p < 0.001) but not sleep fragmentation. Clonidine reduced sleep bout duration and sleep ratio significantly at all three concentrations, and increased sleep fragmentation and velocity significantly at all three concentrations as well as wake bout duration at the highest concentration. Conversely, atomoxetine and guanfacine increased sleep bout duration and sleep ratio significantly at all three concentrations. Sleep fragmentation decreased significantly at all concentrations when treated with atomoxetine but was not affected by guanfacine. Velocity and wake bout duration decreased significantly for all concentrations of guanfacine but only partially for atomoxetine (Supplementary Table 2).
Triage of the Prestwick chemical library
To narrow possible candidates for drug screening in the mutant model, a simple “behavioral fingerprint” [44, 45] for atomoxetine, the most potent compound tested in a prior study [30], was determined in wild-type larvae using sleep parameters. Next, the 1200 compound Prestwick Chemical Library® was screened for matching effects (three concentrations; 16 naïve control groups; 66 DMSO control groups; n = 16; no replicates). The average sleep parameter values were: sleep ratio (59.0 ± 2.32), sleep fragmentation (131.22 ± 4.21), velocity (0.17 ± 0.007 mm/sec), average wake bout duration (10.90 ± 0.39 s) and average sleep bout duration (16.71 ± 1.65 s). Atomoxetine increased fragmentation (up to 145.33 ± 6.12 at the highest dose) and reduced sleep percentage to a range of 29.1–34.4 depending on the dose. Sleep and wake bout durations and velocity were not affected. We thus applied the simplified criterion of a reduction of sleep percentage to less than 40% and increase of fragmentation of 5% to all 1200 compounds.
Aceloclofenac, amlodipine, doxazosin, and moxonidine rescue motility phenotype of adgrl3.1 mutants and interfere with sleep parameters
Sixty-seven compounds met the criterion (above) and were re-assayed using the adgrl3.1−/− mutant larvae (three concentrations; 5 naïve control groups; 5 DMSO control groups; n = 24; no replicates). Four compounds rescued the phenotype.
One-way ANOVA revealed a statistically significant difference between adgrl3.1 control and larvae treated with aceclofenac (F(5, 301) = 12.53, p < 0.001), amlodipine (F(4, 292) = 26.95, p < 0.001), doxazosin (F(5, 304) = 19.28, p < 0.001) and moxonidine (F(5, 321) = 38.97, p < 0.001) for average distance moved (Fig. 3A–D). Dunnett´s post hoc test revealed that moxonidine lowered the average distance moved in a dose dependent manner for 1, 10, 30, and 100 µM doses (Fig. 3D) and amlodipine at 10 and 30 µM doses (Fig. 3B). Aceclofenac lowered the average distance moved for 0.1, 10, and 100 µM doses (Fig. 3A) and doxazosin for all five doses (Fig. 3C) (Supplementary Table 3). The group exposed to 100 µM doxazosin became non-responsive to stimuli a few hours after drug exposure, and 100 µM dose of amlodipine was lethal.
adgrl3.1−/− larvae were treated with five different concentrations (0.1 µM, 1 µM, 10 µM, 30 µM, and 100 µM) of A aceclofenac, B amlodipine, C doxazosin and D moxonidine and compared to larvae treated with 0.3% DMSO. Average distance moved during the five 30-min periods of lights-on and sleep parameters during the night were analyzed. Aceclofenac, amlodipine, doxazosin and moxonidine all reduced the distance moved during lights-on significantly for various concentrations. Aceclofenac, amlodipine, doxazosin and moxonidine all significantly altered the sleep dynamics at multiple parameters during the night. Wild-type (WT) larvae treated with 0.3% DMSO are included for visual comparison. * denote significant differences.
All four drugs did have an effect on the sleep parameters, but to a lesser extent than the non-stimulant ADHD therapeutics tested (Fig. 3). One-way ANOVA revealed that aceclofenac did have a significant effect on the larval sleep fragmentation (F(5,299) = 3.912, p < 0.01) and velocity (F(5,299) = 4.492, p < 0.001) compared to the adgrl3.1−/− homozygous control group, but no significant differences were found for sleep ratio, wake bout duration or sleep bout duration (Fig. 3A). Amlodipine had a significant effect on sleep fragmentation (F(4,288) = 36.845, p < 0.001), sleep ratio (F(4,288) = 4.154, p < 0.01), wake bout duration (F(4,288) = 12.067, p < 0.001) and sleep bout duration (F(4,288) = 3.535, p < 0.01) (Fig. 3B), but no significant differences were found for velocity. Doxazosin did have a significant effect on sleep fragmentation (F(5,301) = 2.426, p < 0.05), sleep ratio (F(5,301) = 6.337, p < 0.001), velocity (F(5,301) = 27.233, p < 0.001) and sleep bout duration (F(5,301) = 3.274, p < 0.01) compared to the adgrl3.1−/− homozygous control group (Fig. 3C), but no significant differences were found for wake bout duration. Lastly, moxonidine had a significant effect on the larvae´s sleep ratio (F(5,306) = 3.325, p < 0.01), velocity (F(5,306) = 10.986, p < 0.001) and sleep bout duration (F(5,306) = 2.53. p < 0.05) (Fig. 3D), but no significant differences were found for sleep fragmentation or wake bout duration. Dunnetts post hoc analysis revealed that significant differences were found between the adgrl3.1−/− homozygous control group and larvae exposed to 0.1 µM and 1 µM aceclofenac, 30 µM amlodipine, 10 µM and 30 µM doxazosin and 10 µM moxonidine for sleep fragmentation. Of these four compounds moxonidine administration resulted in the strongest rescue of the motility phenotype while having the least interference with sleep parameters. Dunnett´s post hoc analysis revealed that the only significant difference for sleep parameters at the dose effective for rescuing the behavioral phenotype, was for sleep fragmentation at 10 µM (Supplementary Table 4).
Moxonidine rescues cognitive deficits in a rodent spontaneous alternation assay
We show that moxonidine also matches the effects of atomoxetine in the spontaneous alternation assay. This is important since this demonstrates efficacy in a different model system and in a different, cognitive, modality. Unpaired t-test revealed that vehicle/scopolamine group showed significant reduction in spontaneous alternations as compared to vehicle/saline/group (67% ± 3 vs. 36% ± 2, t(18) = 7.521, p < 0.001). This decrease in spontaneous alternations reflects cognitive impairment induced by scopolamine. Donepezil and atomoxetine treatment significantly increased (62% ± 2) the spontaneous alternation of scopolamine-mice by 62% ± 2 and 65% ± 3, respectively. Moxonidine treatment resulted in a dose-dependent increase in the spontaneous alternation (41% ± 3, 54% ± 3 and 66% ± 4 for 0.1 mg/kg; 0.3 mg/kg; 1 mg/kg, respectively). The effect was statistically significant (p < 0.001) for the 0.3 and 1 mg/kg doses compared to vehicle (Fig. 4).
A significant difference was found between mice exposed to scopolamine and naïve mice (saline) in which scopolamine-exposed mice showed decrease in spontaneous alterations. Treatment with donepezil and atomoxetine rescued this effect. Moxonidine also rescued this effect in mice in a dose-dependent manner, showing significantly higher spontaneous alterations in mice treated with 0.3 mg/kg and 1 mg/kg treatment but no significant differences were observed between mice treated with 0.1 mg/kg moxonidine compared to scopolamine treated mice. * denote significant differences.
LNP599 rescues the motility phenotype of adgrl3.1 −/− mutants and interferes with sleep parameters
Since moxonidine is a potent I1 agonist, we next assayed a pure I1 receptor agonist, LNP599 [46]. One-way ANOVA revealed a significant difference between adgrl3.1−/− control and LNP599 treated larvae (F(5, 412) = 50.07, p < 0.001) for average distance moved (Fig. 5). Dunnett´s post hoc test revealed that LNP599 lowered the average distance moved in a dose dependent manner (Supplementary Table 5). All sleep parameters, sleep fragmentation (F(5,408) = 5.825, p < 0.001), sleep ratio (F(5,408) = 27.464, p < 0.001), velocity (F(5,408) = 32.004, p < 0.001), wake bout durations (F(5.408) = 7.781, p < 0.001) and sleep bout durations (F(5,408) = 21.474, p < 0.001) were significantly affected. Sleep fragmentation decreased at 30 µM; sleep ratio increased at 10, 30, and 100 µM; velocity decreased at 10, 30, and 100 µM; wake bout durations decreased at 10, 30, and 100 µM and sleep bout durations increased at 30 and 100 µM (Supplementary Table 6).
adgrl3.1−/− larvae were treated with five different concentrations (0.1 µM, 1 µM, 10 µM, 30 µM, and 100 µM) of LNP599 and compared to larvae treated with 0.3% DMSO. Average distance moved during the five 30-min periods of lights-on and sleep parameters during the night were analyzed. LNP599 reduced the distance moved during lights-on significantly in a dose dependent manner for 10 µM, 30 µM, and 100 µM. LNP599 significantly altered all sleep parameters at various concentrations. Wild type (WT) larvae treated with DMSO are included for visual comparison. * denote significant differences.
Discussion
In this study we have characterized a novel adgrl3.1−/− mutant line, and shown that homozygous adgrl3.1−/− mutant larvae are hyperactive compared to their wild-type controls during the light phase. The velocity, however, does not differ during lights-off and neither do their sleep patterns. The model thus fails to capture the sleep disturbances commonly seen in ADHD [47] and, thus leaves no room for therapeutic-driven sleep improvement. However, the model does exhibit the core hyperactive feature of ADHD and is amenable to drug screening [32, 33]. The hyperactive motility phenotype was rescued by atomoxetine, clonidine and guanfacine but with significant impact on the larval sleep profile, consistent with the sleep-promoting properties of α-2 agonists [48] and effects of non-stimulants on sleep in ADHD patients [49]. Four novel compounds rescued the behavioral phenotype of the adgrl3.1−/− mutant model: Aceclofenac, amlodipine, doxasozin and moxonidine. Moxonidine, which had the fewest off-target effects on sleep, was also tested in a rodent model of scopolamine-induced cognitive impairment and was subsequently shown to rescue the deficit to the same extent as both donepezil and atomoxetine, further demonstrating cognitive effects of the compound and establishing similar effects as atomoxetine. Moxonidine, has a 30:1 affinity for I1 receptors over α2 adrenoceptors, which inspired us to test a pure I1 agonist, LNP599 [46] that rescued the behavioral phenotype.
The neural substrates that underpin ADHD are not fully elucidated. However, neuroimaging and functional studies have revealed structural and functional differences between individuals with ADHD and controls [17, 50]. In terms of neurobiology, ADHD is conceptualized as a network disorder encompassing neurochemical dysregulation, in particular at the level of the prefrontal cortex that is involved in executive function and attention [51,52,53]. The anterior cingulate, prefrontal, and orbitofrontal cortices, linked to inattention, hyperactivity, and impulsivity, are interconnected with other cortical and subcortical areas that regulate behavior and emotion. These areas are innervated by DA, NE, and 5-HT projections from the brainstem, and enhanced transmission of these catecholamines represent the major target of ADHD therapeutics [54]. Here we identify potential therapeutics that only partially overlap with known targets.
In general, the current ADHD therapeutics modulate catecholamine neurotransmission, which raises the question about how the novel candidates that rescue the phenotype of the adgrl3.1−/− mutant zebrafish function. Aceclofenac is a non-steroidal anti-inflammatory drug and is a non-selective inhibitor of cyclooxygenase (COX; prostaglandin-endoperoxidase synthase) [55]. COX breaks down arachidonic acid to prostaglandin H2 which in turn is catalyzed to prostaglandin D2 by prostaglandin D synthase (PTGDS) [55, 56]. The PTGDS gene has higher expression in ADHD patients than bipolar patients and lower mRNA expression in bipolar patients than in healthy controls [57, 58]. PTGDS also has a lower expression profile in the spontaneously hypertensive rat (SHR), a classic rodent model of ADHD, compared to control Wistar-Kyoto (WK) rats [59]. Prostaglandin exerts neuromodulatory as well as anti-inflammatory roles and may contribute to the etiology of ADHD, suggesting a role for COX inhibitors, including aceclofenac, in ADHD.
Amlodipine (a dihydropyridine) is an L-type calcium channel (LTCC) blocker [60]. LTCC, Cav1.2 and Cav1.3, encoded by CACNA1C and CACNA1D respectively, are important regulators of calcium influx to neurons and are critical for normal brain development, function and plasticity [61, 62]. CACNA1C and CACNA1D have emerged as highly reproducible candidate risk genes for a variety of neuropsychiatric disorders including ADHD [63]. LTCC´s shape neuronal firing, and are present post-synaptically in signaling complexes where they are poised to regulate activity-dependent transcription by calcium second messenger pathways [64]. Moreover, LTCC´s modulate the release of monoamines and prolong after-hyperpolarization in the prefrontal cortex [64, 65].
Doxasozin (a quinazoline) is a selective α1 adrenergic blocker [66]. Animal studies have revealed that NE binds α2 receptors during alertness whereas it binds α1 under highly stressful conditions suggesting opposite roles of these receptor systems [67, 68]. Excessive NE signaling has been implicated in schizophrenia, PTSD and mania, and α1 antagonism has been suggested as the mechanism of action for many antipsychotics [52, 69]. Studies using the SHR rat have demonstrated that functional α1 receptors persist in juvenile and adult rats whereas they are depleted in control WK rats, resulting in higher spontaneous locus coeruleus (LC) neuronal activity that can be abolished with α1 antagonists [70]. Unsurprisingly, gene enrichment analysis demonstrate enrichment of α1 receptor signaling in ADHD [71].
Moxonidine is a α2 adrenergic as well as I1 receptor agonist [72]. The rescue of the phenotype described here could thus be explained by moxonidine´s α2 agonism. However, moxonidine has an affinity of 33:1 in favor of I1 over α2 [73]. The endogenous ligand of I1 is agmatine, a polyamine synthesized from L-arginine that interacts with 5-HT, cholinergic, α2 and NMDA receptors in addition to I1 [74]. Agmatine has been linked to a variety of beneficial neural effects including neuroprotection, reduction of neuropathic pain and rescue of cognitive symptoms in mice models of Alzheimer´s disease, and a reduction in depressive symptoms as well as amelioration of compulsive-like behaviors [75,76,77,78]. To our knowledge I1 has not been implicated in ADHD previously. Could the behavioral rescue be mediated via the I1 receptor system? Neurobiologically this is plausible via the medial habenula (MH). The main efferent projections of the medial habenula are to the interpeduncular nucleus (IPN) via the fasciculus retroflexus [79, 80]. The IPN in turn gives rise to ascending projections to limbic structures and rodent studies have revealed that the IPN exerts a tonic inhibition on mesocortical, mesolimbic, and mesostriatal dopaminergic neurons [81]. Furthermore, rodent electrophysiological studies have demonstrated strong MH inhibition by agmatine and moxonidine – an effect that is abolished with efaroxan, an I1 antagonist [82]. In zebrafish, habenular lesions increase anxiety-like behaviors and reduce behavioral adaptations [83, 84]. Therefore, it is plausible that I1-mediated habenular inhibition by moxonidine results in less IPN activity, enhancing tonic mesocortical DA transmission which in turn ameliorates the ADHD-like symptoms of the model tested here.
LNP599 is a little-studied pure I1 agonist [46]. The compound has thus far been shown to improve metabolic syndrome, reduce blood pressure and heart rate and reduce plasma catecholamine levels in rodent models [85,86,87,88]. LNP599 also rescues the hyperactive phenotype. The results suggest a role for this receptor system in ADHD that may, at least partially, explain the effects of moxonidine and clonidine.
Before these compounds can be developed into treatments for managing ADHD more assays should be performed including assays for attention and aggression, in addition to verification in a mammalian model. It is noteworthy that three of the compounds have been prescribed for hypertension. Importantly, 14 compounds of the 67-compound subset used for the adgrl3.1−/− assay have been prescribed for hypertension, and nine of them fail to show efficacy in the model, showing that the model is not selective for compounds used to treat hypertension.
In the current paper we describe a novel zebrafish model of ADHD and identify five potential therapeutics for the disease; four repurposed and one novel. The results offer a novel tool to study ADHD, offer insights into the neural substrates of the disease, and identify compounds that could be developed into novel therapeutics.
References
Nigg JT, Sibley MH, Thapar A, Karalunas SL. Development of ADHD: Etiology, heterogeneity, and early life course. Annu Rev Dev Psychol 2020;2:559–83.
Sibley MH, Arnold LE, Swanson JM, Hechtman LT, Kennedy TM, Owens E, et al. Variable patterns of remission from ADHD in the multimodal treatment study of ADHD. Am J Psychiatry. 2022;179:142–51.
Katzman MA, Bilkey TS, Chokka PR, Fallu A, Klassen LJ. Adult ADHD and comorbid disorders: clinical implications of a dimensional approach. BMC Psychiatry. 2017;17:302.
Geffen J, Forster K. Treatment of adult ADHD: a clinical perspective. Ther Adv Psychopharmacol. 2018;8:25–32.
Wu J, Xiao H, Sun H, Zou L, Zhu L-Q. Role of Dopamine receptors in ADHD: A systematic meta-analysis. Mol Neurobiol. 2012;45:605–20.
Nageye F, Cortese S. Beyond stimulants: a systematic review of randomised controlled trials assessing novel compounds for ADHD. Expert Rev Neurother. 2019;19:707–17.
Cortese S. Pharmacologic treatment of attention deficit–hyperactivity disorder. N. Engl J Med. 2020;383:1050–56.
Kooij SJ, Bejerot S, Blackwell A, Caci H, Casas-Brugué M, Carpentier PJ, et al. European consensus statement on diagnosis and treatment of adult ADHD: The European Network Adult ADHD. BMC Psychiatry. 2010;10:67.
Felt BT, Biermann B, Christner JG, Kochhar P, Harrison RV. Diagnosis and management of ADHD in children. Am Fam Phys. 2014;90:456–64.
Briars L, Todd T. A review of pharmacological management of attention-deficit/hyperactivity disorder. J Pediatr Pharmacol Ther. 2016;21:192–206.
Carolan D. ADHD stimulant medication misuse and considerations for current prescribing practice: a literature review. Ir J Med Sci. 2022;191:313–32.
Mechler K, Banaschewski T, Hohmann S, Häge A. Evidence-based pharmacological treatment options for ADHD in children and adolescents. Pharmacol Ther. 2021;230:107940.
Nasser A, Liranso T, Adewole T, Fry N, Hull JT, Busse GD, et al. A Phase 3, Placebo-controlled trial of once-daily viloxazine extended-release capsules in adolescents with attention-deficit/hyperactivity disorder. J Clin Psychopharmacol. 2021;41:370–80.
Pang L, Sareen R. Retrospective analysis of adverse events associated with non-stimulant ADHD medications reported to the united states food and drug administration. Psychiatry Res. 2021;300:113861.
Mamiya PC, Arnett AB, Stein MA. Precision medicine care in ADHD: The case for neural excitation and inhibition. Brain Sci. 2021;13;11:91.
Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2019;24:562–75.
Yadav SK, Bhat AA, Hashem S, Nisar S, Kamal M, Syed N, et al. Genetic variations influence brain changes in patients with attention-deficit hyperactivity disorder. Transl Psychiatry. 2021;11:349.
Li Z, Chang SH, Zhang LY, Gao L, Wang J. Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry Res 2014;219:10–24.
Arcos-Burgos M, Velez JI, Martinez AF, Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, et al. ADGRL3 (LPHN3) variants predict substance use disorder. Transl Psychiatry. 2019;9:42.
Bruxel EM, Moreira-Maia CR, Akutagava-Martins GC, Quinn TP, Klein M, Franke B, et al. Meta-analysis and systematic review of ADGRL3 (LPHN3) polymorphisms in ADHD susceptibility. Mol Psychiatry. 2021;26:2277–85.
Huang X, Zhang Q, Gu X, Hou Y, Wang M, Chen X, et al. LPHN3 gene variations and susceptibility to ADHD in Chinese Han population: a two-stage case-control association study and gene-environment interactions. Eur Child Adolesc Psychiatry. 2019;28:861–73.
Mastronardi CA, Pillai E, Pineda DA, Martinez AF, Lopera F, Velez JI, et al. Linkage and association analysis of ADHD endophenotypes in extended and multigenerational pedigrees from a genetic isolate. Mol Psychiatry. 2016;21:1434–40.
Sando R, Jiang X, Sudhof TC. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science. 2019;363:eaav7969.
Regan SL, Hufgard JR, Pitzer EM, Sugimoto C, Hu Y-C, Williams MT, et al. Knockout of latrophilin-3 in Sprague-Dawley rats causes hyperactivity, hyper-reactivity, under-response to amphetamine, and disrupted dopamine markers. Neurobiol Dis. 2019;130:104494.
Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–66.
Ribasés M, Ramos-Quiroga JA, Sánchez-Mora C, Bosch R, Richarte V, Palomar G, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes, Brain Behav. 2011;10:149–57.
Fallgatter AJ, Ehlis AC, Dresler T, Reif A, Jacob CP, Arcos-Burgos M, et al. Influence of a latrophilin 3 (LPHN3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (ADHD). Eur Neuropsychopharmacol.2013;23:458–68.
Regan SL, Pitzer EM, Hufgard JR, Sugimoto C, Williams MT, Vorhees CV. A novel role for the ADHD risk gene latrophilin-3 in learning and memory in Lphn3 knockout rats. Neurobiol Dis. 2021;158:105456.
Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ, et al. Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res. 2012;1463:85–92.
Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, et al. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry. 2012;17:946–54.
Regan SL, Williams MT, Vorhees CV. Review of rodent models of attention deficit hyperactivity disorder. Neurosci Biobehav Rev. 2021;132:621–37.
Fontana BD, Norton WHJ, Parker MO. Modelling ADHD-like phenotypes in Zebrafish. Curr Top Behav Neurosci. 2022;57:395–414.
Fontana BD, Franscescon F, Rosemberg DB, Norton WHJ, Kalueff AV, Parker MO. Zebrafish models for attention deficit hyperactivity disorder (ADHD). Neurosci Biobehav Rev. 2019;100:9–18.
Lavalou P, Eckert H, Damy L, Constanty F, Majello S, Bitetti A, et al. Strategies for genetic inactivation of long noncoding RNAs in Zebrafish. RNA 2019;25:897–904.
Sveinsdottir HS, Decker A, Christensen C, Lucena PB, Thornorsteinsson H, Richert E, et al. Motility phenotype in a zebrafish vmat2 mutant. PLoS One. 2022;17:e0259753.
Christensen C, Þorsteinsson H, Maier VH, Karlsson KAE. Multi-parameter behavioral phenotyping of the MPP+ Model of Parkinson’s disease in Zebrafish. Front Behav Neurosci. 2020;14:623924.
Barlow IL, Rihel J. Zebrafish sleep: from geneZZZ to neuronZZZ. Curr Opin Neurobiol. 2017;44:65–71.
Sorribes A, Thornorsteinsson H, Arnardottir H, Johannesdottir I, Sigurgeirsson B, de Polavieja GG, et al. The ontogeny of sleep-wake cycles in zebrafish: a comparison to humans. Front Neural Circuits. 2013;7:178.
Sigurgeirsson B, Thornorsteinsson H, Sigmundsdottir S, Lieder R, Sveinsdottir HS, Sigurjonsson OE, et al. Sleep-wake dynamics under extended light and extended dark conditions in adult zebrafish. Behav Brain Res. 2013;256:377–90.
Niijima-Yaoita F, Nagasawa Y, Tsuchiya M, Arai Y, Tadano T, Tan-No K. Effects of methylphenidate on the impairment of spontaneous alternation behavior in mice intermittently deprived of REM sleep. Neurochem Int. 2016;100:128–37.
de Bruin NM, Prickaerts J, Lange JH, Akkerman S, Andriambeloson E, de Haan M, et al. SLV330, a cannabinoid CB1 receptor antagonist, ameliorates deficits in the T-maze, object recognition and social recognition tasks in rodents. Neurobiol Learn Mem. 2010;93:522–31.
Maelicke A, Hoeffle-Maas A, Ludwig J, Maus A, Samochocki M, Jordis U, et al. Memogain is a galantamine pro-drug having dramatically reduced adverse effects and enhanced efficacy. J Mol Neurosci. 2010;40:135–7.
Andriambeloson E, Huyard B, Poiraud E, Wagner S. Methyllycaconitine- and scopolamine-induced cognitive dysfunction: differential reversal effect by cognition-enhancing drugs. Pharm Res Perspect. 2014;2:e00048.
Rihel J, Schier AF. Behavioral screening for neuroactive drugs in zebrafish. Dev Neurobiol. 2012;72:373–85.
Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010;327:348–51.
Gasparik V, Greney H, Schann S, Feldman J, Fellmann L, Ehrhardt JD, et al. Synthesis and biological evaluation of 2-aryliminopyrrolidines as selective ligands for I1 imidazoline receptors: discovery of new sympatho-inhibitory hypotensive agents with potential beneficial effects in metabolic syndrome. J Med Chem. 2015;58:878–87.
Becker SP. ADHD and sleep: recent advances and future directions. Curr Opin Psychol. 2020;34:50–56.
Weerink MAS, Struys M, Hannivoort LN, Barends CRM, Absalom AR, Colin P. Clinical pharmacokinetics and pharmacodynamics of Dexmedetomidine. Clin Pharmacokinet. 2017;56:893–913.
Coogan AN, Schenk M, Palm D, Uzoni A, Grube J, Tsang AH, et al. Impact of adult attention deficit hyperactivity disorder and medication status on sleep/wake behavior and molecular circadian rhythms. Neuropsychopharmacology 2019;44:1198–206.
Saad JF, Griffiths KR, Korgaonkar MS. A systematic review of imaging studies in the combined and inattentive subtypes of attention deficit hyperactivity disorder. Front Integr Neurosci. 2020;14:31.
Del Campo N, Chamberlain SR, Sahakian BJ, Robbins TW. The roles of dopamine and noradrenaline in the pathophysiology and treatment of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2011;69:e145–57.
Arnsten AF, Rubia K. Neurobiological circuits regulating attention, cognitive control, motivation, and emotion: disruptions in neurodevelopmental psychiatric disorders. J Am Acad Child Adolesc Psychiatry. 2012;51:356–67.
Kasparek T, Theiner P, Filova A. Neurobiology of ADHD from childhood to adulthood: findings of imaging methods. J Atten Disord. 2015;19:931–43.
Sharma A, Couture J. A review of the pathophysiology, etiology, and treatment of attention-deficit hyperactivity disorder (ADHD). Ann Pharmacother. 2014;48:209–25.
Iolascon G, Gimenez S, Mogyorosi D. A review of Aceclofenac: Analgesic and anti-inflammatory effects on musculoskeletal disorders. J Pain Res. 2021;14:3651–63.
Ambati GG, Jachak SM. Natural product inhibitors of Cyclooxygenase (COX) enzyme: a review on current status and future perspectives. Curr Med Chem. 2021;28:1877–905.
Marin-Mendez JJ, Patino-Garcia A, Segura V, Ortuno F, Galvez MD, Soutullo CA. Differential expression of prostaglandin D2 synthase (PTGDS) in patients with attention deficit-hyperactivity disorder and bipolar disorder. J Affect Disord. 2012;138:479–84.
Munkholm K, Peijs L, Kessing LV, Vinberg M. Reduced mRNA expression of PTGDS in peripheral blood mononuclear cells of rapid-cycling bipolar disorder patients compared with healthy control subjects. Int J Neuropsychopharmacol. 2014;18:5.
Qiu J, Hong Q, Chen RH, Tong ML, Zhang M, Fei L, et al. Gene expression profiles in the prefrontal cortex of SHR rats by cDNA microarrays. Mol Biol Rep. 2010;37:1733–40.
van Zwieten PA. Amlodipine: an overview of its pharmacodynamic and pharmacokinetic properties. Clin Cardiol. 1994;17:III3–6.
Kabir ZD, Martinez-Rivera A, Rajadhyaksha AM. From gene to behavior: L-Type Calcium channel mechanisms underlying neuropsychiatric symptoms. Neurotherapeutics 2017;14:588–613.
Kabir ZD, Lee AS, Rajadhyaksha AM. L-type Ca(2+) channels in mood, cognition and addiction: integrating human and rodent studies with a focus on behavioural endophenotypes. J Physiol. 2016;594:5823–37.
Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013;381:1371–79.
Stanford SC. Psychostimulants, antidepressants and neurokinin-1 receptor antagonists (‘motor disinhibitors’) have overlapping, but distinct, effects on monoamine transmission: the involvement of L-type Ca2+ channels and implications for the treatment of ADHD. Neuropharmacology 2014;87:9–18.
Zamponi GW, Striessnig J, Koschak A, Dolphin AC. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharm Rev. 2015;67:821–70.
Young RA, Brogden RN. Doxazosin. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy in mild or moderate hypertension. Drugs 1988;35:525–41.
Arnsten AF, Pliszka SR. Catecholamine influences on prefrontal cortical function: relevance to treatment of attention deficit/hyperactivity disorder and related disorders. Pharm Biochem Behav. 2011;99:211–6.
Akinaga J, Garcia-Sainz JA, S Pupo A. Updates in the function and regulation of alpha1 -adrenoceptors. Br J Pharmacol 2019;176:2343–57.
Perez DM. alpha1-Adrenergic receptors in neurotransmission, synaptic plasticity, and cognition. Front Pharmacol. 2020;11:581098.
Igata S, Hayashi T, Itoh M, Akasu T, Takano M, Ishimatsu M. Persistent alpha1-adrenergic receptor function in the nucleus locus coeruleus causes hyperexcitability in AD/HD model rats. J Neurophysiol. 2014;111:777–86.
Hayman V, Fernandez TV. Genetic insights into ADHD biology. Front Psychiatry. 2018;9:251.
Ernsberger P. Pharmacology of moxonidine: an I1-imidazoline receptor agonist. J Cardiovasc Pharmacol. 2000;35:S27–41.
Bousquet P, Hudson A, Garcia-Sevilla JA, Li JX. Imidazoline receptor system: the past, the present, and the future. Pharm Rev. 2020;72:50–79.
Neis VB, Rosa PB, Olescowicz G, Rodrigues ALS. Therapeutic potential of agmatine for CNS disorders. Neurochem Int. 2017;108:318–31.
Aizawa H, Zhu M. Toward an understanding of the habenula’s various roles in human depression. Psychiatry Clin Neurosci. 2019;73:607–12.
Kotagale N, Deshmukh R, Dixit M, Fating R, Umekar M, Taksande B. Agmatine ameliorates manifestation of depression-like behavior and hippocampal neuroinflammation in mouse model of Alzheimer’s disease. Brain Res Bull. 2020;160:56–64.
Kotagale N, Dixit M, Garmelwar H, Bhondekar S, Umekar M, Taksande B. Agmatine reverses memory deficits induced by Abeta1-42 peptide in mice: A key role of imidazoline receptors. Pharm Biochem Behav. 2020;196:172976.
Kotagale NR, Taksande BG, Inamdar NN. Neuroprotective offerings by agmatine. Neurotoxicology 2019;73:228–45.
Andres KH, von During M, Veh RW. Subnuclear organization of the rat habenular complexes. J Comp Neurol. 1999;407:130–50.
McLaughlin I, Dani JA, De, Biasi M. The medial habenula and interpeduncular nucleus circuitry is critical in addiction, anxiety, and mood regulation. J Neurochem. 2017;142:130–43.
Nishikawa T, Fage D, Scatton B. Evidence for, and nature of, the tonic inhibitory influence of habenulointerpeduncular pathways upon cerebral dopaminergic transmission in the rat. Brain Res. 1986;373:324–36.
Weiss T, Bernard R, Bernstein HG, Veh RW, Laube G. Agmatine modulates spontaneous activity in neurons of the rat medial habenular complex-a relevant mechanism in the pathophysiology and treatment of depression? Transl Psychiatry. 2018;8:201.
Agetsuma M, Aizawa H, Aoki T, Nakayama R, Takahoko M, Goto M, et al. The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nat Neurosci. 2010;13:1354–6.
Mathuru AS, Jesuthasan S. The medial habenula as a regulator of anxiety in adult zebrafish. Front Neural Circuits. 2013;7:99.
Estato V, Nascimento A, Antunes B, Gomes F, Coelho L, Rangel R, et al. Cerebral microvascular dysfunction and inflammation are improved by centrally acting antihypertensive drugs in metabolic syndrome. Metab Syndr Relat Disord. 2017;15:26–35.
Nascimento AR, Gomes F, Machado MV, Goncalves-de-Albuquerque C, Bousquet P, Tibirica E. I1-imidazoline receptor-mediated cardiovascular and metabolic effects in high-fat diet-induced metabolic syndrome in rats. Auton Neurosci. 2019;217:18–25.
Fellmann L, Regnault V, Greney H, Gasparik V, Muscat A, Max JP, et al. A new pyrroline compound selective for I1-imidazoline receptors improves metabolic syndrome in rats. J Pharm Exp Ther. 2013;346:370–80.
Weiss M, Fellmann L, Regnard P, Bousquet P, Monassier L, Niederhoffer N. Protective effects of the imidazoline-like drug lnp599 in a marmoset model of obesity-induced metabolic disorders. Int J Obes. 2021;45:1229–39.
Funding
The study was funded in full by 3Z.
Author information
Authors and Affiliations
Contributions
KÆK and HÞ designed the study. PL and AS generated the mutated strain. HÞ, HSS, and CC collected and analyzed zebrafish data and edited the manuscript. EA and SW collected and analyzed mouse data and edited the manuscript. WHJN, MP, CC, and HÞ revised manuscript. KÆK wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Authors Hildur Sóley Sveinsdóttir, Christian Christensen, Perrine Lavalou, Matthew O. Parker, Alena Shkumatava, William H.J. Norton, Emile Andriambeloson and Stéphanie Wagner declare no biomedical financial interests or potential conflicts of interest. Karl Ægir Karlsson and Haraldur Þorsteinsson are co-founders and shareholders in 3Z which funded the study in full.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sveinsdóttir, H.S., Christensen, C., Þorsteinsson, H. et al. Novel non-stimulants rescue hyperactive phenotype in an adgrl3.1 mutant zebrafish model of ADHD. Neuropsychopharmacol. 48, 1155–1163 (2023). https://doi.org/10.1038/s41386-022-01505-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-022-01505-z