Abstract
Animal and human studies have documented the existence of developmental windows (or sensitive periods) when experience can have lasting effects on brain structure or function, behavior, and disease. Although sensitive periods for depression likely arise through a complex interplay of genes and experience, this possibility has not yet been explored in humans. We examined the effect of genetic pathways regulating sensitive periods, alone and in interaction with common childhood adversities, on depression risk. Guided by a translational approach, we: (1) performed association analyses of three gene sets (60 genes) shown in animal studies to regulate sensitive periods using summary data from a genome-wide association study of depression (n = 807,553); (2) evaluated the developmental expression patterns of these genes using data from BrainSpan (n = 31), a transcriptional atlas of postmortem brain samples; and (3) tested gene-by-development interplay (dGxE) by analyzing the combined effect of common variants in sensitive period genes and time-varying exposure to two types of childhood adversity within a population-based birth cohort (n = 6254). The gene set regulating sensitive period opening associated with increased depression risk. Notably, 6 of the 15 genes in this set showed developmentally regulated gene-level expression. We also identified a statistical interaction between caregiver physical or emotional abuse during ages 1–5 years and genetic risk for depression conferred by the opening genes. Genes involved in regulating sensitive periods are differentially expressed across the life course and may be implicated in depression vulnerability. Our findings about gene-by-development interplay motivate further research in large, more diverse samples to further unravel the complexity of depression etiology through a sensitive period lens.
Similar content being viewed by others
Introduction
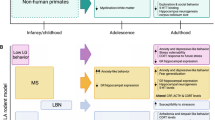
Sensitive periods are stages of heightened plasticity when experience can have particularly strong and enduring effects on brain structure, behavior, and health [1,2,3,4,5]. To date, sensitive periods have been most commonly studied with respect to sensory systems and related domains, including vision, hearing, and language learning, in animals [6,7,8] and humans [9, 10]. This research has revealed that sensitive period plasticity occurs through an orchestration of genes and life experiences [11, 12]. As shown in Fig. 1 and previously summarized elsewhere [12], robust evidence from in vivo experiments with genetically modified mice or rats has shown that several dozen genes regulate the opening, closing, and expression of sensitive periods in the visual and auditory systems [12,13,14,15]. For instance, some opening genes (e.g., Bdnf or Gad2 [12, 16, 17]) initiate, accelerate, or delay the onset of sensitive periods by regulating parvalbumin (PV) cell maturation and altering the ratio of excitatory and inhibitory circuit activity. Some closing genes (e.g., Acan and Rtn4r) can regulate the formation of perineuronal nets (PNNs), which operate as a “molecular brake” of sensitive period plasticity [18]. Expression genes (e.g., Nr2a or Stat1 [19, 20]) maintain the duration of sensitive periods by circuit rewiring and consolidation. Beyond the primary sensory cortex, these genetic pathways have been recently implicated in plasticity mechanisms that configure the prefrontal cortical network [21], which regulates cognition and mood [8, 18, 21]. Thus, alterations in genetic pathways involved in sensitive period regulation have developmental impacts across brain regions and could give rise to varying levels of psychiatric vulnerability.
Gene sets identified by Hensch [13] and Takesian and Hensch [12]. Opening genes: BDNF, ARNTL, CLOCK, GABBR1, GABRA1, GABRA2, GABRB3, GAD1, GAD2, SLC6A1, NTRK2, OTX2, NPTX2, HTR3A, CHRNA4. Closing genes: OTX2, GCLC, GCLM, LYNX1, MAG, MBP, HLA-C, HLA-A, HLA-B, PODN, LILRB3, LILRB1, PTPRS, RTN4, ACAN, BCAN, HAPLN1, HAPLN3, HAPLN4, NCAN, PTS, TNR, VCAN, ADAMTS15, ADAMTS4, ADAMTS8, CSGALNACT1, HAS1, HAS2, HAS3, MME, MMP15, MMP24, MMP3, MMP8, DLG4, PILRB, CAM5, NCAM1. Expression genes: CREB1, KCNK2, NGF, GRIN2A, DLG4, PVALB, STAT1, TNF. (Note: 60 genes are presented in total; OTX2 and DLG4 are duplicated).
To that end, accumulating evidence from molecular studies suggest genetic dysregulation of sensitive period plasticity may explain risk for neuropsychiatric disorders. For instance, in mouse models of autism, researchers observed mistimed sensitive period onset due to premature or impoverished PV circuits [22, 23]. In both clinical patients and animal models, reduction of sensitive period triggers (PV + interneurons) or molecular brakes (PNNs) and sensitive period-related transcriptional or epigenetic aberrations confer greater risk for schizophrenia [22,23,24]. Recent findings from large-scale genome-wide association studies (GWAS) also point towards associations between variants located in genes implicated in sensitive period regulation (e.g., GABBR1 [22, 23, 25, 26], GRIN2A [24, 27,28,29,30], NCAM1 [26, 31], and NCAN [30]) and neuropsychiatric illnesses such as schizophrenia, depression, and bipolar disorder.
Environmental perturbations during sensitive periods are also associated with risk for neuropsychiatric disorders [32, 33]. Observational epidemiologic studies in humans show that exposure to childhood adversity may have time-dependent effects on brain structure and function [34], as well as social, emotional, and behavioral processes, ranging from fear conditioning to stress reactivity and psychopathology symptoms [4, 35, 36]. This work has generally found that adversity during the first 5 years of life is associated with the greatest risk for psychiatric outcomes relative to exposure after age 5 years or no exposure [4]. Adversity is thought to disrupt sensitive period functioning through experience-expectant processes, wherein the brain is primed through genetic instruction during restricted periods of development to expect a normative set of environmental inputs [37]. Experiences of childhood adversity, including acts of social commission (e.g., physical or sexual abuse) or social and material omission (e.g., neglect, poverty) [38]) are therefore understood as violations of expected environmental inputs that can lead to impaired brain plasticity and mental disorders [39].
Depression is thought to be governed by the interplay between genetic variations and time-dependent experiences over the life course [40, 41]. Although the timing and duration of depression-related sensitive periods are unknown, preclinical and molecular studies have implicated sensitive period biology in the etiology of depression. For example, the molecular signature of sensitive period closure, PNNs, has protective effects against oxidative stress [42]. Oxidative stress responses are connected to the pathophysiology of major depressive disorder (MDD) [43, 44] and are impacted by childhood adversity [45, 46]. Moreover, deficits in GABAergic transmission, which plays a key role in regulating sensitive period timing, can alter susceptibility to early-life stress [47]. Therefore, stress exposures during developmental periods of heightened vulnerability may give rise to more severe symptoms of depression, especially among individuals with genetic variation linked to disrupted sensitive period timing [47,48,49].
However, to our knowledge, no studies have investigated the independent and joint roles of gene sets regulating developmental plasticity and exposure to time-varying early-life adversity on depression risk. Here we pursued the overarching hypothesis that genetic variation governing sensitive period plasticity interacts with adverse life experiences during specific developmental windows to shape risk for depression. We tested this hypothesis using a translational approach, bridging and triangulating sources of evidence (from animal models and human studies) [50], types of data (cross-sectional and longitudinal), and disciplines (genetics, developmental neuroscience, and epidemiology). Adopting a pathway-based approach similar to previous studies on neuropsychiatric disorder etiology [51, 52], we focused on three sets of genes encompassing existing molecular evidence for sensitive period biology, which regulate the opening (n = 15 genes), closing (n = 39 genes), and expression (n = 8 genes) of sensitive periods (conceptual framework shown in Fig. 1). These genes were identified from a thorough review of preclinical literature on genetic variation involved in regulating developmental plasticity. Given the predominance of evidence from sensory cortex, we focused mainly on these brain regions while reviewing the literature, although similar mechanisms have been identified in subcortical structures [53,54,55]. The following three research questions are sequentially investigated (as shown in Fig. 2):
-
(1)
Does variation in sensitive period genes identified from animal models predict risk for depression in humans?
-
(2)
Are the sensitive period genes implicated in depression risk developmentally regulated? If so, can we identify variants in these genes that function as developmental expression quantitative trait loci (which we refer to as “d-QTLs”), shaping developmental timing of expression patterns in the brain?
-
(3)
Does variation in sensitive period genes interact with time-dependent effects of adversity to shape levels of depressive symptoms? In other words, is there evidence for developmental gene–environment interplay (dG × E)?
We approached the above three questions systematically and sequentially, with results from each stage of analysis guiding our approach at the next stage. Specifically, gene sets associated with depression risk in question 1 analysis were used to address question 2, where we characterized the developmental trajectories of their gene expression levels and examined whether the trajectories were shaped by certain variants. Similarly, variants annotated to genes in gene sets associated with depression risk were used to compute a gene-set genetic risk score in question 3. Results from question 2 analysis suggested the existence of a biologically defined sensitive period based on levels of gene expression, which guided how the timing of environmental exposure was parameterized in question 3 analysis.
Methods and materials
Research question 1: examining the association between sensitive period gene sets and risk for depression
Genetic data
To evaluate the strength of association between genetic pathways involved in regulating sensitive period functioning and risk for depression, we analyzed summary statistics from a GWAS meta-analysis of depression, comprising 414,055 cases and 892,299 controls from 3 large-scale depression samples: UK Biobank, the Psychiatric Genomics Consortium, and 23andMe, Inc. [56]. Across subsamples, depression was defined using minimal (e.g., self-reported symptoms) and deep phenotyping (e.g., structured clinical interviews) approaches. Therefore, the analyses captured genetic architecture of general depression, rather than strict clinical diagnosis of MDD. Details about the genotyping and quality-control procedure are provided by Howard et al. [56].
Data analysis
We pursued a pathway analysis of summary data from the meta-analysis described above. Specifically, we performed competitive gene-set analyses using the Multi-marker Analysis of GenoMic Annotation (MAGMA) software (Version 1.06) [57]. MAGMA uses a nested approach to first summarize single-nucleotide polymorphism (SNP)-level associations into gene-level associations and then gene set-level associations, allowing for the detection of aggregated signals at even modest levels. In the current analysis, SNP-level estimates were obtained from the meta-analysis of depression described above [56]. For the gene-level analyses, we annotated each of the SNPs reported in the summary data to genes using human genome build 37 (hg19) as the reference. Gene-level p-values were calculated using the sum of −log(p-values) of all tested SNPs within a gene, defined as the region between the transcription start and stop sites of each gene [57]. For the gene set-level analyses, we grouped the 60 sensitive period genes into their respective sets based on their biological functions (Fig. 1). To perform a competitive gene-set analysis, we compared the gene-level results for our three sensitive period gene sets to the gene-level results of the rest of the genome; this test determined whether the average association between genes in the given set and depression was stronger than that of other genes not in the set. We included a corrected p-value that empirically accounted for multiple testing, using 10,000 random permutations. In contrast to single genetic variant-level analyses, this gene-set approach tested specific biological pathways to assess their potential as therapeutic targets [57]. Moreover, in the context of polygenicity (i.e., thousands of loci each conferring modest risk), aggregating functionally consistent signals can substantially improve statistical power [40, 58].
Of note, five genes in our analysis were located in the major histocompatibility complex (MHC), a region of the genome involved in human immunity that contains polymorphic loci with long-range linkage-disequilibrium (LD) patterns [59]. Although these features could make interpretation of SNP-level associations more difficult, we did not remove SNPs in the extended MHC region, as LD is explicitly modeled in MAGMA to produce unbiased results (see Supplemental Materials).
Research question 2: investigating the developmental regulation of depression-implicated sensitive period gene sets
Gene expression data
We investigated the temporal expression patterns of sensitive period genes within the left hemisphere of three brain regions involved in the pathophysiology of depression (amygdala, hippocampus, and medial prefrontal cortex (mPFC)) [60]. Data came from BrainSpan [61, 62] (http://www.brainspan.org), a transcriptional atlas of healthy, postmortem brain donors (ages 5.7 weeks post conception to 82 years) without large-scale genomic or other abnormalities (see Supplemental Materials). In brief, the 31 postnatal donors studied represented both sexes (42% were female) and were predominantly of European ancestry (58%). BrainSpan investigators classified donors by developmental stage at time of death: 0–5 months (n = 3); 6–11 months (n = 3); 1–5 years (n = 2); 6–11 years (n = 3); 12–19 years (n = 4); 20–39 years (n = 9); 40–59 years (n = 4); and 60+ years (n = 3). Prenatal donors and the right hemisphere data were not analyzed here, due to lack of data suitability or availability (see Supplemental Materials).
Quality-controlled, quantile-normalized exon microarray data [63] were downloaded from Gene Expression Omnibus (GSE25219). For each brain region, expression levels for all probes within an exon were averaged to obtain an expression value for each exon. Probes were annotated to genes using the UCSC Human Genome (hg19) reference sequence. Similar to prior studies [64, 65], we used the median of all exons within each gene as the estimate of gene expression. Expression values are presented in log(2) values; thus, each one-unit difference represents a doubling of expression. Although RNA-sequencing data were available in BrainSpan, that dataset contained only one-third of the sample size, which was too small for our analysis (see Supplemental Materials).
Data analysis
To investigate whether the expression patterns of sensitive period genes were developmentally regulated, we tested the hypothesis that developmental stage at the time of death explained a significant amount of variation in gene expression. Specifically, we performed multiple regression analysis using an omnibus F-test to compare the full model (with developmental stage included as a categorical variable) to a baseline model (only adjusting for two principal components capturing genetic ancestry). The amount of variation in gene expression additionally explained by developmental stage was quantified using the increase in R2. To reduce multiple testing burden, we focused only on genes in the gene sets associated with depression risk from analyses of the first research question.
Further, to explore whether certain genetic variants could shape developmental expression patterns of sensitive period genes implicated in depression, we tested the interactions between genotype and developmental stage on expression. Genotype was included as a categorical variable instead of the conventional additive model, to capture the potential nonlinear relationship between genotype and trajectories of gene expression; developmental stage was modeled as an ordinal variable, with both linear and quadratic effects to account for nonlinearity while retaining model parsimony. An F-test was then performed to compare the genotype-by-developmental timing model (with the interaction terms) to the baseline model (without the interaction terms). Of note, we restricted these exploratory analyses to the mPFC, as we saw the most evidence for developmental regulation in this brain region. More details are provided in Supplemental Materials.
Research question 3: investigating interactions between genome-wide and gene set-level genetic liability to depression, timing of exposure to adversity, and depressive symptoms in development (i.e., dG × E)
Dataset and measures
To examine potential dG × E, we analyzed data from 6254 child participants in the Avon Longitudinal Study of Parents and Children (ALSPAC), a UK-based prospective, longitudinal birth cohort of children followed for more than two decades [66,67,68]. Approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Consent for biological samples was collected in accordance with the Human Tissue Act 2004 [69]. More details are available on the ALSPAC website, including a fully searchable data dictionary: http://www.bristol.ac.uk/alspac/researchers/our-data/.
The analytic sample included all participants who were genotyped and had data on the outcome, average depressive symptoms from 10.5 to 23 years of age [70, 71]. Details about the genotype data collection and quality-control procedure are provided in Supplemental Materials. Depressive symptoms across adolescence and into young adulthood were measured by averaging clinically administered and child self-reports of depressive symptoms using the Short Mood and Feelings Questionnaire [72] (Supplemental Materials). We focused on average symptoms to maximize the analytic sample size and measure general levels of symptoms across development.
We studied two commonly occurring and frequently studied types of adversity, both of which have been consistently associated with psychopathology symptoms in childhood [73] and adulthood [74, 75]. They represent examples of how expectations of environmental inputs may be disrupted during development [76, 77]. Prior literature [73, 78] and our own analyses (see Supplemental Materials) provided converging evidence of a time-dependent relationship between exposures to socioeconomic disadvantage or caregiver physical or emotional abuse and depressive symptoms. Socioeconomic disadvantage was measured as a time-varying construct based on maternal reports of the extent to which the family had difficulty affording items for the child, rent or mortgage, heating, clothing, or food. Similarly, caregiver physical or emotional abuse was measured through repeated assessments completed by the mother and her partner, which reported whether either of them had been physically or emotionally cruel to the child. Exposure was defined as a three-level variable: no exposure before age 10.5 years, exposure during ages 1–5 years, and exposure at other time points outside the sensitive period (grouped into before or after the sensitive period).
Data analysis
Using summary statistics provided by Howard et al. [56] as weights, we generated a polygenic risk score (PRS) representing risk for depression conferred by common variants in the gene set(s) associated with depression in analyses of the first research question (3617 SNPs before clumping). As a comparison, we additionally generated a genome-wide PRS including all SNPs associated with depression in the summary statistics at the threshold of p < 0.05. Prior to computing the PRS, clumping was performed to eliminate SNPs that were in high LD (r2 > 0.25). PRS calculations were conducted in PLINK 1.90 [79].
All analyses controlled for the following covariates (measured at childbirth): sex, maternal age, number of previous pregnancies, home ownership, highest level of maternal education, and maternal marital status. We also adjusted for the top four genetic principal components to control for population stratification. To reduce potential bias and maximize statistical power, missing exposure and covariate data were multiply-imputed using the MICE package [80] in R among participants with data on average depressive symptoms. All subsequent multiple regression analyses were performed using 20 imputed datasets. Estimates were combined to account for variation between- and within-imputed datasets. To examine whether patterns of developmental gene-by-environment interplay differed between male and female participants, we performed sex-stratified analyses as a secondary investigation.
Results
Are genetic pathways involved in regulating sensitive periods associated with depression risk?
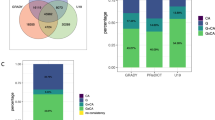
Of the three gene sets examined, only the gene set regulating the opening of sensitive periods was associated with risk for depression (corrected p-value = 0.01, Table 1). There was no evidence for associations between gene sets involved in the closing or expression of sensitive periods and depression risk.
Are sensitive period genes implicated in depression risk developmentally regulated?
Developmental stage was significantly associated with expression levels in six opening genes, explaining up to 54% of the additional variation in gene-level expression beyond genetic ancestry (Supplemental Table S1). We observed evidence for developmental regulation in the mPFC but not hippocampus or amygdala (Fig. 3). Three of the six opening genes with evidence of developmental regulation had a nadir of expression between ages 1 and 5 years: gene expression levels between ages 1 and 5 years were significantly different from other time points at GABRA1 (β = −2.33, 95% confidence interval (95% CI) [−3.47, −1.19], p = 3 × 10−4), GAD1 (β = −1.88, 95% CI [−2.78, −0.98], p = 2 × 10−4), and GAD2 (β = −2.32, 95% CI [−3.25, −1.39], p = 2 × 10−5).
AMY, amygdala; HIP, hippocampus; mPFC, medial prefrontal cortex. Thicker lines indicated a significant association between developmental stage and gene expression (i.e., developmental regulation) based on results from multiple regression analyses. The evidence for developmental regulation was found in the mPFC. In particular, three genes showed decreased expression between ages 1 and 5 years (GABRA1, GAD1, and GAD2).
We further examined whether genotype was associated with developmental expression of these genes. To identify SNPs associated with developmental gene expression (d-QTLs), we tested SNP-by-age interactions on expression levels of the opening genes. Analyses were limited to the mPFC and examined 144 independent SNPs (Supplemental Materials).
We found nominal evidence (p < 0.05) for SNP-by-age interactions at five loci (Supplemental Table S2), with two SNPs (rs1442060, an intron variant in GABRA2; rs7900976, an intron variant in GAD2) showing significant associations after accounting for the number of SNPs tested within each gene. GABRA2 was upregulated early in life and downregulated later in life for both major and minor allele homozygotes, whereas the expression level was more stable over time for heterozygotes (Supplemental Fig. S1). GAD2 was upregulated over time for major allele homozygotes and downregulated for heterozygous individuals (Supplemental Fig. S1). There were only two data points available from homozygous minor individuals, so the trend over time was not discernible. These findings suggest that genetic variants may be associated with different patterns of gene expression over the life course.
How does variation in sensitive period genes interact with the timing of exposure to adversity to shape depressive symptoms?
Average depressive symptoms across adolescence were heritable in our sample (h2SNP = 9.1%, SE = 0.05, p = 0.03). The genome-wide PRS for depression was significantly associated with average depressive symptoms (β = 0.40, 95% CI [0.30, 0.49], p < 1 × 10−22).
The gene set-level PRS representing all SNPs annotated to the opening genes did not have a main effect on the outcome (β = −0.01, 95% CI [−0.11, 0.08], p = 0.76). Socioeconomic disadvantage showed a time-dependent association with depressive symptoms. Specifically, exposure between ages 1 and 5 years was associated with increased symptoms (β = 0.84, 95% CI [0.57, 1.11], p = 7.86 × 10−10). This effect estimate was twice as large as the increase associated with exposure at other time points (β = 0.37, 95% CI [−0.01, 0.74], p = 0.05). Exposure to caregiver physical or emotional abuse during both time periods, meaning between ages 1–5 years or at other time points outside this range, had similar associations with increased depressive symptoms (βages1–5 = 0.87, 95% CI [0.54, 1.20], p = 3.08 × 10−7; βother = 0.83, 95% CI [0.47, 1.19], p = 6.23 × 10−6; Supplemental Tables S5 and S6).
When assessing the main genetic and environmental effects additively, we observed similar patterns: both the genome-wide PRS and childhood adversity exposures were associated with depressive symptoms, and no effect of gene set-level PRS was detected (Fig. 3, Models 4 and 5). Although there was no evidence supporting dG × E for socioeconomic disadvantage (Fig. 3, Models 6 and 7), we found evidence for dG × E effects for caregiver physical or emotional abuse. Specifically, the opening gene PRS was unassociated with depressive symptoms among participants reporting no exposure or exposure at time points outside the sensitive period, whereas a higher opening gene PRS was associated with higher levels of depressive symptoms among participants exposed between ages 1 and 5 years (Fig. 4, Model 7). These findings suggest that signals for gene-by-development interplay may be adversity and pathway specific (Fig. 5).
Depressive symptoms were measured using the Short Mood and Feelings Questionnaire, which ranged on a scale of 0–26, as represented on the Y-axis. The opening genetic score summarizes genetic variations associated with depression in a set of genes involved in regulating the opening of sensitive periods, based on primarily preclinical studies of cortical regions of the brain. The intercepts and slopes plotted above were based on fitted regression models examining potential gene-by-development interplay using data from the ALSPAC (Fig. 4, Model 7). A Lines representing a linear relationship between the opening genetic score and depressive symptoms in different exposure groups did not show any obvious patterns of cross-over. B We observed evidence for a statistical interaction, where a positive association between the genetic score and depressive symptoms was only observed for individuals exposed to caregiver physical or emotional abuse between ages 1 and 5 years in B.
In testing for sex-specific effects, we also observed opposing effect estimates for opening gene risk scores and the effects of childhood adversity on depressive symptoms, which revealed a general pattern of females being at greater risk than males (Supplemental Materials).
Discussion
Three notable findings emerged from our analyses. First, we found that genetic variation in genes governing the onset of sensitive periods was associated with risk for depression at the population level. Although the mechanism underlying this association remains unknown, we posit that, because these genes regulate the initial maturation of inhibitory signaling during early development [6, 12], their altered function might therefore delay the onset of sensitive period plasticity. Such effects could result in aberrant responses to external stimuli during the process of fear learning and other cognitive functions critical to affective development [81], and thus increase the risk for depression and other neuropsychiatric disorders. Notably, OTX2, one of the genes involved in sensitive period onset, mediates the effect of early-life stress on brain structures implicated in depression and lasting susceptibility to stress both in animal models and in humans [82, 83].
By contrast, we found no effect of genetic pathways involved in the duration or closure of sensitive periods on depression. The lack of association with closing genes is somewhat surprising given previous studies suggesting that PNN maturation plays an instrumental role in the emergence of higher-order functioning (e.g., social memory formation) [84] and has protective effects against toxic stress exposures [42]. The lack of evidence from our study could be due to the heterogeneity among genes included in the closing set. Among the 39 genes in this set, some relate to the formation of PNNs (e.g., ACAN, RTN4R), whereas others affect myelin and myelin-associated inhibitors that restrict plasticity (e.g., PIRB) [12]. Although these 39 genes all regulate the closure of sensitive periods, they had mixed effects on depression risk. Namely, only 11 out of the 39 genes showed gene-level associations with depression risk (Supplemental Table S7 and Fig. S2). We analyzed the entire closing set to preserve the biological interpretation of this pathway and avoid cherry-picking; however, testing somewhat heterogeneous genes as one pathway might have diluted the signals.
Second, we showed that genes involved in sensitive period functioning were developmentally regulated in the mPFC, although not the hippocampus or amygdala. Configurations of the prefrontal cortical network during sensitive periods have been implicated in different developmental trajectories and vulnerability to neuropsychiatric disorders, suggesting that plasticity of mPFC may play a prominent role in development [21]. The absence of evidence for developmental regulation in the amygdala or hippocampus was unexpected, because the trajectories of GABAergic signaling markers in these regions vary over time in animal models [85]. Our inability to identify time-varying patterns in these regions could be attributed to the limited sample size; although they were not significant, some genes (e.g., GABRA1, GAD1, and GAD2) did show similar trends of expression in the amygdala and hippocampus. The lack of discernable developmental regulation might also be due to variability in inhibitory mechanisms across subregions of brain structures. For example, the dorsal and ventral mPFC have opposing effects on fear expression and extinction, limiting the ability to detect differences when analyzing the mPFC as a whole [86, 87]. We were unable to tease apart substructure differences in the current study, as subregion data were unavailable.
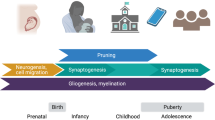
Moreover, the nadir in expression levels of opening genes between ages 1 and 5 years suggests that the regulation of these genes is particularly relevant during this time period. This decline in the transcription of plasticity-related genes co-occurred with reported periods of decreased synaptogenesis and increased pruning or maturation of the central nervous system, which may signal the start of developing higher-order cognition or more complex behavior [88, 89]. Although previous literature has pointed to heightened sensitivity to stress in mPFC during adolescence due to dysregulated glucocorticoid production [90], our results point to an earlier period of developmental sensitivity marked by the downregulation of GABA signaling pathways. However, we were unable to fully disentangle different patterns between individuals across time due to the lack of repeated gene expression measures. Future studies are needed to confirm the importance of the developmental window between ages 1 and 5 years for the regulation of these opening genes. We also provided a proof of concept for identifying genetic loci predicting developmental regulation of genes—which we call d-QTLs. Although our results are preliminary given the restrictions of sample size and data availability, the d-QTL framework holds potential for studying the heterogeneity of sensitive period timing and responsivity across individuals with different genetic profiles.
Third, in a population-based sample of adolescents, we did not observe any main effects of genetic risk for depression conferred by the opening gene-set pathway. However, we found evidence for its interaction with exposure to caregiver physical or emotional abuse during a potential sensitive period, ages 1–5 years. This gene-by-developmental interplay suggests that variation in the genetic regulation of sensitive period onsets may exert little or no influence on depression risk in children receiving typical environmental inputs. However, when coupled with a stressful environment during specific periods of development, a higher genetic predisposition toward dysregulated sensitive periods could lead to heightened vulnerabilities [91]. As this interaction was only observed for one type of childhood adversity, the mechanisms underlying the biological embedding of exposures and psychopathology could be adversity specific. Socioeconomic disadvantage captures elements of both material deprivation and heightened psychosocial stress. Exposure in early life, especially during a sensitive period of plasticity, could have a lasting impact on development through structural alterations in the social environment [92, 93] or other biological mechanisms, including epigenetic regulation or genetically orchestrated neural mechanisms such as glucocorticoid dysregulation, which may ultimately alter individual responses to early-life stress [90, 92]. In addition, our data came from a homogeneous sample of European ancestry children in the United Kingdom, where most families of participants were relatively socioeconomically advantaged [67]. Thus, we were unable to capture large detrimental effects of extreme financial hardship and their potential interactions with genetic variation.
Several limitations of our study are noted. First, we focused on the analysis of sensitive period-regulating genes that were first identified from rodent studies of primary sensory cortical structures. Nevertheless, similar sensitive period mechanisms are implicated in the plasticity of more evolutionarily conserved subcortical structures [94], such as the amygdala and hippocampus. For example, Gogolla et al. [53] showed that formation of PNNs coincided with a developmental critical period of fear extinction in the amygdala. If and when other key genetic components of developmental plasticity in subcortical regions emerge, future population-based studies should further examine those pathways. Second, the lack of detailed phenotypic data in BrainSpan made it impossible for us to test whether the developmental regulation of genetic pathways was impacted by life experiences, and whether different trajectories may contribute to depression risk. By studying associations between phenotypic experiences and gene expression levels across time in humans, future studies can better link observable environmental risks to underlying molecular changes. Further, the limited sample size in BrainSpan reduced the generalizability and power of our analyses, which should be addressed when larger databases of brain gene expression across the life span become available. Third, measures of depression varied across research questions: whereas depression examined in Question 1 (from the GWAS of depression) was a minimally phenotyped binary construct, depressive symptoms were measured through research-clinic administered and self-reported questionnaires in Question 3 (ALSPAC). Although a minimal phenotype may still yield important biological insights in large samples [95], discrepancies between phenotype operationalizations should be kept in mind as we draw connections across datasets. Fourth, all three data sources comprised predominantly White populations, restricting the generalizability of our findings. As African American and non-White Hispanic adolescents experience a greater burden of childhood adversity [96, 97], interactions between genetic risk and environmental exposures may also disproportionally contribute to susceptibility for mental health disorders in racial and ethnic minority populations and perpetuate health disparities [98]. It should remain a priority of neuropsychiatric genetic studies to expand the recruitment and research of non-White samples [99]. Larger individual studies or meta/mega-analyses including high-risk samples of marginalized communities and rigorous assessments of phenotypes are also warranted.
In conclusion, our study provides a translational model for testing novel neurobiological pathways, triangulating concepts and data from genetics, developmental sciences, and epidemiology. Through the integration of genetic risk, experiences over the life course, and physiological markers across domains of functioning, interdisciplinary translational studies hold great potential to unravel the complexity of depression etiology and identify novel targets for prevention or intervention.
Funding and disclosure
Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award Numbers K01MH102403 and R01MH113930 (ECD) and K24MH094614 (JWS). It was also supported in part by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (ECD) and an award from the Jacobs Foundation (ECD and AET). The Psychiatric Genomics Consortium (PGC) has received major funding from the US National Institute of Mental Health and the US National Institute of Drug Abuse (U01 MH109528 and U01 MH1095320). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). GWAS data were generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe, Inc. This publication is the work of the authors and all authors will serve as guarantors for the contents of this paper. JWS is an unpaid member of the Bipolar/Depression Research Community Advisory Panel of 23andMe, Inc., a member of the Leon Levy Foundation Neuroscience Advisory Board, and has received an honorarium for an internal seminar at Biogen, Inc. He is the Principal Investigator of a collaborative study of the genetics of depression and bipolar disorder sponsored by 23andMe, Inc., for which 23andMe, Inc. provides analysis time as in-kind support but no payments. The remaining authors (including the contributing Consortium members) have nothing to disclose.
References
Bornstein MH. Sensitive periods in development: structural characteristics and causal interpretations. Psychol Bull. 1989;105:179–97.
Bailey DB, Bruer JT, Symons FJ, Lichtman JW. Critical thinking about critical periods. A series from the National Center for Early Development and Learning (Baltimore: Brookes Publishing, 2001).
Knudsen EI. Sensitive periods in the development of the brain and behavior. J Cogn Neurosci. 2004;16:1412–25.
Dunn EC, McLaughlin KA, Slopen N, Rosand J, Smoller JW. Developmental timing of child maltreatment and symptoms of depression and suicidal ideation in young adulthood: Results from the National Longitudinal Study on Adolescent Health. Depress Anxiety. 2013;30:10.
Uliana DL, Gomes FV, Grace AA. Stress impacts corticoamygdalar connectivity in an age-dependent manner. Neuropsychopharmacology. 2021;46:731–40.
Hensch TK. Critical period regulation. Annu Rev Neurosci. 2004;27:549–79.
Hooks BM, Chen C. Critical periods in the visual system: changing views for a model of experience-dependent plasticity. Neuron. 2007;56:312–26.
Yang E-J, Lin EW, Hensch TK. Critical period for acoustic preference in mice. Proc Natl Acad Sci USA. 2012;109:17213–20.
Sharma A, Campbell J, Cardon G. Developmental and cross-modal plasticity in deafness: Evidence from the P1 and N1 event related potentials in cochlear implanted children. Int J Psychophysiol. 2015;95:135–44.
Hakuta K, Bialystok E, Wiley E. Critical evidence: a test of the critical-period hypothesis for second-language acquisition. Psychol Sci. 2003;14:31–8.
Fox SE, Levitt P, Nelson CA. How the timing and quality of early experiences influence the development of brain architecture. Child Dev. 2010;81:28–40.
Takesian AE, Hensch TK. Balancing plasticity/stability across brain development. Prog Brain Res. 2013;207:3–34.
Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–88.
Kaneko M, Stellwagen D, Malenka RC, Stryker MP. Tumor necrosis factor-α mediates one component of competitive, experience-dependent plasticity in developing visual cortex. Neuron. 2008;58:673–80.
Tropea D, Van Wart A, Sur M. Molecular mechanisms of experience-dependent plasticity in visual cortex. Philos Trans R Soc Lond B Biol Sci. 2009;364:341–55.
Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, et al. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–55.
Anomal R, de Villers-Sidani E, Merzenich MM, Panizzutti R. Manipulation of BDNF signaling modifies the experience-dependent plasticity induced by pure tone exposure during the critical period in the primary auditory cortex. PLoS ONE. 2013;8:e64208.
Lee HHC, Bernard C, Ye Z, Acampora D, Simeone A, Prochiantz A, et al. Genetic Otx2 mis-localization delays critical period plasticity across brain regions. Mol Psychiatry. 2017;22:680–8.
Fagiolini M, Katagiri H, Miyamoto H, Mori H, Grant SGN, Mishina M, et al. Separable features of visual cortical plasticity revealed by N-methyl-d-aspartate receptor 2A signaling. Proc Natl Acad Sci USA. 2003;100:2854–9.
Nagakura I, Wart AV, Petravicz J, Tropea D, Sur M. STAT1 regulates the homeostatic component of visual cortical plasticity via an AMPA receptor-mediated mechanism. J Neurosci. 2014;34:10256–63.
Guirado R, Perez-Rando M, Ferragud A, Gutierrez-Castellanos N, Umemori J, Carceller H, et al. A critical period for prefrontal network configurations underlying psychiatric disorders and addiction. Front Behav Neurosci. 2020;14:51.
Hall LS, Adams MJ, Arnau-Soler A, Clarke T-K, Howard DM, Zeng Y, et al. Genome-wide meta-analyses of stratified depression in Generation Scotland and UK Biobank. Transl Psychiatry. 2018;8:9.
Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44.
Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–83.
Yu H, Yan H, Li J, Li Z, Zhang X, Ma Y, et al. Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol Psychiatry. 2017;22:954–60.
Nagel M, Jansen PR, Stringer S, Watanabe K, Leeuw dCA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Lam M, Hill WD, Trampush JW, Yu J, Knowles E, Davies G, et al. Pleiotropic meta-analysis of cognition, education, and schizophrenia differentiates roles of early neurodevelopmental and adult synaptic pathways. Am J Hum Genet. 2019;105:334–50.
Periyasamy S, John S, Padmavati R, Rajendren P, Thirunavukkarasu P, Gratten J, et al. Association of schizophrenia risk with disordered niacin metabolism in an Indian genome-wide association study. JAMA Psychiatry. 2019;76:1026–34.
Ikeda M, Takahashi A, Kamatani Y, Momozawa Y, Saito T, Kondo K, et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr Bull. 2019;45:824–34.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Baselmans BML, Jansen R, Ip HF, Dongen VJ, Abdellaoui A, Weijer VDMP, et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet. 2019;51:445–51.
Johnson FK, Delpech J-C, Thompson GJ, Wei L, Hao J, Herman P, et al. Amygdala hyper-connectivity in a mouse model of unpredictable early life stress. Transl Psychiatry. 2018;8:1–14.
Peña CJ, Smith M, Ramakrishnan A, Cates HM, Bagot RC, Kronman HG, et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat Commun. 2019;10:5098.
Andersen SL, Tomada A, Vincow ES, Valente E, Polcari A, Teicher MH. Preliminary evidence for sensitive periods in the effect of childhood sexual abuse on regional brain development. J Neuropsychiatry Clin Neurosci. 2008;20:292–301.
Bos K, Zeanah CH, Fox NA, Drury SS, McLaughlin KA, Nelson CA. Psychiatric outcomes in young children with a history of institutionalization. Harv Rev Psychiatry. 2011;19:15–24.
Dunn EC, Nishimi K, Powers A, Bradley B. Is developmental timing of trauma exposure associated with depressive and post-traumatic stress disorder symptoms in adulthood? J Psychiatr Res. 2017;84:119–27.
Greenough WT, Black JE, Wallace CS. Experience and brain development. Child Dev. 1987;58:539–59.
Gilbert R, Widom CS, Browne K, Fergusson D, Webb E, Janson S. Burden and consequences of child maltreatment in high-income countries. Lancet. 2009;373:68–81.
McLaughlin KA, Sheridan MA, Nelson CA. Neglect as a violation of species-expectant experience: neurodevelopmental consequences. Biol Psychiatry. 2017;82:462–71.
Dunn EC, Brown RC, Dai Y, Rosand J, Nugent NR, Amstadter AB, et al. Genetic determinants of depression: recent findings and future directions. Harv Rev Psychiatry. 2015;23:1–18.
Uher R, Zwicker A. Etiology in psychiatry: embracing the reality of poly-gene-environmental causation of mental illness. World Psychiatry. 2017;16:121–9.
Do KQ, Cuenod M, Hensch TK. Targeting oxidative stress and aberrant critical period plasticity in the developmental trajectory to schizophrenia. Schizophr Bull. 2015;41:835–46.
Black CN, Bot M, Scheffer PG, Cuijpers P, Penninx BWJH. Is depression associated with increased oxidative stress? A systematic review and meta-analysis. Psychoneuroendocrinology. 2015;51:164–75.
Liu T, Zhong S, Liao X, Chen J, He T, Lai S, et al. A meta-analysis of oxidative stress markers in depression. PLoS ONE. 2015;10:e0138904.
Theall KP, Drury SS, Shirtcliff EA. Cumulative neighborhood risk of psychosocial stress and allostatic load in adolescents. Am J Epidemiol. 2012;176:S164–S74.
Drury SS, Theall K, Gleason MM, Smyke AT, De Vivo I, Wong JYY, et al. Telomere length and early severe social deprivation: linking early adversity and cellular aging. Mol Psychiatry. 2012;17:719–27.
Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406.
Cameron JL, Eagleson KL, Fox NA, Hensch TK, Levitt P. Social origins of developmental risk for mental and physical illness. J Neurosci. 2017;37:10783–91.
LeMoult J, Humphreys KL, Tracy A, Hoffmeister J-A, Ip E, Gotlib IH. Meta-analysis: exposure to early life stress and risk for depression in childhood and adolescence. J Am Acad Child Adolesc Psychiatry. 2020;59:842–55.
Bale TL, Abel T, Akil H, Carlezon Jr WA, Moghaddam B, Nestler EJ, et al. The critical importance of basic animal research for neuropsychiatric disorders. Neuropsychopharmacology. 2019;44:1349–53.
The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Rammos A, Gonzalez LAN, Weinberger DR, Mitchell KJ, Nicodemus KK. The role of polygenic risk score gene-set analysis in the context of the omnigenic model of schizophrenia. Neuropsychopharmacology. 2019;44:1562–9.
Mota NR, Poelmans G, Klein M, Torrico B, Fernàndez-Castillo N, Cormand B, et al. Cross-disorder genetic analyses implicate dopaminergic signaling as a biological link between attention-deficit/hyperactivity disorder and obesity measures. Neuropsychopharmacology. 2020;45:1188–95.
Gogolla N, Caroni P, Lüthi A, Herry C. Perineuronal nets protect fear memories from erasure. Science. 2009;325:1258–61.
Lybrand ZR, Goswami S, Zhu J, Jarzabek V, Merlock N, Aktar M, et al. A critical period of neuronal activity results in aberrant neurogenesis rewiring hippocampal circuitry in a mouse model of epilepsy. Nat Commun. 2021;12:1423.
Tenkumo C, Ohta K-I, Suzuki S, Warita K, Irie K, Teradaya S, et al. Repeated maternal separation causes transient reduction in BDNF expression in the medial prefrontal cortex during early brain development, affecting inhibitory neuron development. Heliyon. 2020;6:e04781.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–54.
Glanville KP, Coleman JRI, Hanscombe KB, Euesden J, Choi SW, Purves KL, et al. Classical human leukocyte antigen alleles and C4 haplotypes are not significantly associated with depression. Biol Psychiatry. 2020;87:419–30.
Heim C, Binder EB. Current research trends in early life stress and depression: Review of human studies on sensitive periods, gene–environment interactions, and epigenetics. Exp Neurol. 2012;233:102–11.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9.
Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–29.
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Das D, Clark TA, Schweitzer A, Yamamoto M, Marr H, Arribere J, et al. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35:4845–57.
Laiho A, Elo LL. A note on an exon-based strategy to identify differentially expressed genes in RNA-Seq experiments. PLoS ONE. 2014;9:e115964.
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort profile: The ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–27.
Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort profile: The Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110.
Northstone K, Lewcock M, Groom A, Boyd A, Macleod J, Timpson N, et al. The Avon Longitudinal Study of Parents and Children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51.
Human Tissue Act (2004).
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Informatics. 2009;42:377–81.
Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Informatics. 2019;95:103208.
Messer SC, Angold A, Costello J, Loeber R, Van Kammen W, Stouthamer-Loeber M. Development of a short questionnaire for use in epidemiological studies of depression in children and adolescents: factor composition and structure across development. Int J Methods Psychiatric Res. 1995;5:251–62.
Dunn EC, Soare TW, Raffeld MR, Busso DS, Crawford KM, Davis KA, et al. What life course theoretical models best explain the relationship between exposure to childhood adversity and psychopathology symptoms: recency, accumulation, or sensitive periods? Psychol Med. 2018;48:2562–72.
McLaughlin KA, Breslau J, Green JG, Lakoma MD, Sampson NA, Zaslavsky AM, et al. Childhood socio-economic status and the onset, persistence, and severity of DSM-IV mental disorders in a US national sample. Social Sci Med. 2011;73:1088–96.
Boe T, Balaj M, Eikemo TA, McNamara CL, Solheim EF. Financial difficulties in childhood and adult depression in Europe. Eur J Public Health. 2017;27:96–101.
Slopen N, Koenen KC, Kubzansky LD. Childhood adversity and immune and inflammatory biomarkers associated with cardiovascular risk in youth: a systematic review. Brain Behav Immunity. 2012;26:239–50.
Slopen N, Koenen KC, Kubzansky LD. Cumulative adversity in childhood and emergent risk factors for long-term health. J Pediatrics. 2014;164:631–38.e2.
Teicher MH, Anderson CM, Polcari A. Childhood maltreatment is associated with reduced volume in the hippocampal subfields CA3, dentate gyrus, and subiculum. Proc Natl Acad Sci USA. 2012;109:E563–E72.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira Manuel AR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw. 2011;45:1548–7660.
Hartley CA, Lee FS. Sensitive periods in affective development: nonlinear maturation of fear learning. Neuropsychopharmacology. 2015;40:50–60.
Peña CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–8.
Kaufman J, Wymbs NF, Montalvo-Ortiz JL, Orr C, Albaugh MD, Althoff R, et al. Methylation in OTX2 and related genes, maltreatment, and depression in children. Neuropsychopharmacology. 2018;43:2204–11.
Domínguez S, Rey CC, Therreau L, Fanton A, Massotte D, Verret L, et al. Maturation of PNN and ErbB4 signaling in area CA2 during adolescence underlies the emergence of PV interneuron plasticity and social memory. Cell Rep. 2019;29:1099–112.e4.
King EC, Pattwell SS, Sun A, Glatt CE, Lee FS. Nonlinear developmental trajectory of fear learning and memory. Ann N Y Acad Sci. 2013;1304:62–9.
Adhikari A, Lerner TN, Finkelstein J, Pak S, Jennings JH, Davidson TJ, et al. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–85.
Sierra-Mercado D, Padilla-Coreano N, Quirk GJ. Dissociable roles of prelimbic and infralimbic cortices, ventral hippocampus, and basolateral amygdala in the expression and extinction of conditioned fear. Neuropsychopharmacology. 2011;36:529–38.
Huttenlocher PR, de Courten C, Garey LJ, Van der Loos H. Synaptogenesis in human visual cortex — evidence for synapse elimination during normal development. Neurosci Lett. 1982;33:247–52.
Silbereis John C, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89:248–68.
Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10:434–45.
Dunn EC, Uddin M, Subramanian SV, Smoller JW, Galea S, Koenen KC. Research review: gene–environment interaction (GxE) research in youth depression – a systematic review with recommendations for future research. J Child Psychol Psychiatry Allied Disciplines. 2011;52:1223–38.
Cerutti JK, Lussier AA, Zhu Y, Liu J, Dunn EC. Associations between indicators of socioeconomic position and DNA methylation: A systematic review. medRxiv:2021:2021.01.21.21250199 [Preprint]. 2021. Available from: https://doi.org/10.1101/2021.01.21.21250199.
Poulton R, Caspi A, Milne BJ, Thomson WM, Taylor A, Sears MR, et al. Association between children’s experience of socioeconomic disadvantage and adult health: a life-course study. Lancet. 2002;360:1640–5.
Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019;51:1624–36.
Cai N, Revez JA, Adams MJ, Andlauer TFM, Breen G, Byrne EM, et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat Genetics. 2020;52:437–47.
Ahern J, Karasek D, Luedtke AR, Bruckner TA, van der Laan MJ. Racial/ethnic differences in the role of childhood adversities for mental disorders among a nationally representative sample of adolescents. Epidemiology. 2016;27:697–704.
Slopen N, Shonkoff JP, Albert MA, Yoshikawa H, Jacobs A, Stoltz R, et al. Racial disparities in child adversity in the U.S.: interactions with family immigration history and income. Am J Preventive Med. 2016;50:47–56.
Dunn EC, Wiste A, Radmanesh F, Almli LM, Gogarten SM, Sofer T, et al. Genome-wide association study (GWAS) and genome-wide environment interaction study (GWEIS) of depressive symptoms in African American and Hispanic/Latina women. Depress Anxiety. 2016;33:265–80.
Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–8.
Acknowledgements
We thank Oliver Hoffman, PhD, formerly of the Harvard Chan Bioinformatics Core, Harvard TH Chan School of Public Health, Boston, MA, Meeta Mistry, Virginia Fisher, and Kuang Zheng for their assistance with early data analyses. We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. We also thank the research participants and employees of 23andMe, Inc. for making this work possible.
Author information
Authors and Affiliations
Consortia
Contributions
YZ, JWS, TKH, and ECD conceived and designed the analysis. ECD and contributing authors of the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium acquired the data. YZ, MJW, KMC, JCRT, and KAD performed the analysis. AAL, CdL, AET, TKH, JWS, and ECD provided critical feedback on preliminary results and revised the analysis. YZ, MJW, KMC, and ECD drafted the manuscript. All authors critically reviewed the manuscript, gave their final approval, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Corresponding authors
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhu, Y., Wang, MJ., Crawford, K.M. et al. Sensitive period-regulating genetic pathways and exposure to adversity shape risk for depression. Neuropsychopharmacol. 47, 497–506 (2022). https://doi.org/10.1038/s41386-021-01172-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41386-021-01172-6
This article is cited by
-
ASMT determines gut microbiota and increases neurobehavioral adaptability to exercise in female mice
Communications Biology (2023)