Abstract
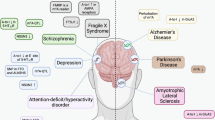
Various chemical modifications of all RNA transcripts, or epitranscriptomics, have emerged as crucial regulators of RNA metabolism, attracting significant interest from both basic and clinical researchers due to their diverse functions in biological processes and immense clinical potential as highlighted by the recent profound success of RNA modifications in improving COVID-19 mRNA vaccines. Rapid accumulation of evidence underscores the critical involvement of various RNA modifications in governing normal neural development and brain functions as well as pathogenesis of brain disorders. Here we provide an overview of RNA modifications and recent advancements in epitranscriptomic studies utilizing animal models to elucidate important roles of RNA modifications in regulating mammalian neurogenesis, gliogenesis, synaptic formation, and brain function. Moreover, we emphasize the pivotal involvement of RNA modifications and their regulators in the pathogenesis of various human brain disorders, encompassing neurodevelopmental disorders, brain tumors, psychiatric and neurodegenerative disorders. Furthermore, we discuss potential translational opportunities afforded by RNA modifications in combatting brain disorders, including their use as biomarkers, in the development of drugs or gene therapies targeting epitranscriptomic pathways, and in applications for mRNA-based vaccines and therapies. We also address current limitations and challenges hindering the widespread clinical application of epitranscriptomic research, along with the improvements necessary for future progress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Crick F. Central dogma of molecular biology. Nature. 1970;227:561–3.
Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18:275–91.
Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42.
Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119–31.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24.
Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74.
Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552–9.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187–1200.
Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–9.
Morais P, Adachi H, Yu YT. The Critical Contribution of Pseudouridine to mRNA COVID-19 Vaccines. Front Cell Dev Biol. 2021;9:789427.
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–53.
Vissers C, Sinha A, Ming GL, Song H. The epitranscriptome in stem cell biology and neural development. Neurobiol Dis. 2020;146:105139.
Shafik AM, Allen EG, Jin P. Dynamic N6-methyladenosine RNA methylation in brain and diseases. Epigenomics. 2020;12:371–80.
Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21:36–51.
Shafik AM, Allen EG, Jin P. Epitranscriptomic dynamics in brain development and disease. Mol Psychiatry. 2022;27:3633–46.
Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell. 2017;171:877–89 e817.
Bohnsack KE, Hobartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m(5)C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes. 2019;10:102.
He PC, He C. m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 2021;40:e105977.
Kumar S, Mohapatra T. Deciphering Epitranscriptome: Modification of mRNA Bases Provides a New Perspective for Post-transcriptional Regulation of Gene Expression. Front Cell Dev Biol. 2021;9:628415.
Nombela P, Miguel-Lopez B, Blanco S. The role of m(6)A, m(5)C and Psi RNA modifications in cancer: novel therapeutic opportunities. Mol cancer. 2021;20:18.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46.
Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72.
Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. M6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat Methods. 2016;13:692–8.
Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458.
Li X, Zhu P, Ma S, Song J, Bai J, Sun F, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592–7.
Carlile TM, Rojas-Duran MF, Gilbert WV. Pseudo-Seq: Genome-Wide Detection of Pseudouridine Modifications in RNA. Methods Enzymol. 2015;560:219.
Meyer KD. DART-seq: an antibody-free method for global m6A detection. Nat Methods. 2019;16:1275–80.
Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–56.
Mirza AH, Attarwala N, Gross SS, Chen Q, Jaffrey SR. Selective detection of m6A derived from mRNA using the Phospho-tag m6A assay. bioRxiv. 2022;493172.
Ensinck I, Sideri T, Modic M, et al. m6A-ELISA, a simple method for quantifying N6-methyladenosine from mRNA populations. RNA. 2023;29:705–12.
Richard EM, Polla DL, Assir MZ, Contreras M, Shahzad M, Khan AA, et al. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am J Hum Genet. 2019;105:869–78.
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–8.
Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41.
Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP Modulates Neural Differentiation through m(6)A-Dependent mRNA Nuclear Export. Cell Rep. 2019;28:845–54.e845.
Martinez NM, Su A, Burns MC, Nussbacher JK, Schaening C, Sathe S, et al. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol Cell. 2022;82:645–59.e649.
Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204.
Borchardt EK, Martinez NM, Gilbert WV. Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet. 2020;54:309–36.
Li X, Ma S, Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol. 2016;33:108–16.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639.
Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an mC reader. Cell Res. 2017;27:606–25.
Kleiber N, Lemus-Diaz N, Stiller C, Heinrichs M, Mai MM, Hackert P, et al. The RNA methyltransferase METTL8 installs m(3)C(32) in mitochondrial tRNAs(Thr/Ser(UCN)) to optimise tRNA structure and mitochondrial translation. Nat Commun. 2022;13:209.
Scholler E, Marks J, Marchand V, Bruckmann A, Powell CA, Reichold M, et al. Balancing of mitochondrial translation through METTL8-mediated m(3)C modification of mitochondrial tRNAs. Mol Cell. 2021;81:4810–25.e4812.
Zhang F, Yoon K, Zhang DY, Kim NS, Ming GL, Song H. Epitranscriptomic regulation of cortical neurogenesis via Mettl8-dependent mitochondrial tRNA m(3)C modification. Cell Stem Cell. 2023;30:300–11.e311.
Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, et al. Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem. 2017;292:14695–703.
Ignatova VV, Kaiser S, Ho JSY, Bing X, Stolz P, Tan YX, et al. METTL6 is a tRNA m(3)C methyltransferase that regulates pluripotency and tumor cell growth. Sci Adv. 2020;6:eaaz4551.
Shima H, Igarashi K. N 1-methyladenosine (m1A) RNA modification: the key to ribosome control. J Biochem. 2020;167:535–9.
Zhang C, Jia G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteom Bioinforma. 2018;16:155–61.
Dimitrova DG, Teysset L, Carré C. RNA 2'-O-Methylation (Nm) Modification in Human Diseases. Genes. 2019;10:117.
Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. RNA ribose methylation (2’-O-methylation): Occurrence, biosynthesis and biological functions. Biochim Biophys Acta Gene Regul Mech. 2019;1862:253–69.
Oerum S, Meynier V, Catala M, Tisne C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239–55.
Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X, et al. Landscape and Regulation of m(6)A and m(6)Am Methylome across Human and Mouse Tissues. Mol Cell. 2020;77:426–40.e426.
Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, et al. Identification of the m(6)Am Methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol Cell. 2019;75:631–43 e638.
Cowling VH. Regulation of mRNA cap methylation. Biochem J. 2009;425:295–302.
Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–26.
Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019;74:1278–90.e1279.
Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–66.
Cheng W, Gao A, Lin H, Zhang W. Novel roles of METTL1/WDR4 in tumor via m(7)G methylation. Mol Ther Oncolytics. 2022;26:27–34.
Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 2019;29:927–41.
Chalk AM, Taylor S, Heraud-Farlow JE, Walkley CR. The majority of A-to-I RNA editing is not required for mammalian homeostasis. Genome Biol. 2019;20:268.
Wang H, Chen S, Wei J, Song G, Zhao Y. A-to-I RNA Editing in Cancer: From Evaluating the Editing Level to Exploring the Editing Effects. Front Oncol. 2020;10:632187.
Torres AG, Pineyro D, Filonava L, Stracker TH, Batlle E, Ribas de Pouplana L. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 2014;588:4279–86.
Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021;184:3109–24.e3122.
Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021;22:375–92.
Zhang LS, Xiong QP, Pena Perez S, Liu C, Wei J, Le C, et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat Cell Biol. 2021;23:684–91.
Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–14.
Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–57.
Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19:45–58.
Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16:98–112.
Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal Chem. 2018;90:6380–4.
Drazkowska K, Tomecki R, Warminski M, Baran N, Cysewski D, Depaix A, et al. 2’-O-Methylation of the second transcribed nucleotide within the mRNA 5’ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 2022;50:9051–71.
Cirzi C, Dyckow J, Legrand C, Schott J, Guo W, Perez Hernandez D, et al. Queuosine-tRNA promotes sex-dependent learning and memory formation by maintaining codon-biased translation elongation speed. EMBO J. 2023;42:e112507.
Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin cell Biol. 2005;17:648–57.
Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43.
Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206.
Du K, Zhang Z, Zeng Z, Tang J, Lee T, Sun T. Distinct roles of Fto and Mettl3 in controlling development of the cerebral cortex through transcriptional and translational regulations. Cell Death Dis. 2021;12:700.
Niu F, Che P, Yang Z, Zhang J, Yang L, Zhuang M, et al. m(6)A regulation of cortical and retinal neurogenesis is mediated by the redundant m(6)A readers YTHDFs. iScience. 2022;25:104908.
Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69.
Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68.
Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Rep. 2017;8:112–24.
Chen P, Zhang T, Yuan Z, Shen B, Chen L. Expression of the RNA methyltransferase Nsun5 is essential for developing cerebral cortex. Mol Brain. 2019;12:74.
Cheng IC, Chen BC, Shuai HH, Chien FC, Chen P, Hsieh TS. Wuho Is a New Member in Maintaining Genome Stability through its Interaction with Flap Endonuclease 1. PLoS Biol. 2016;14:e1002349.
Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–50.
Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702.
Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X, et al. m(6)A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genomics Proteom Bioinforma. 2019;17:154–68.
Sun W, Zhang B, Bie Q, Ma N, Liu N, Shao Z. The Role of RNA Methylation in Regulating Stem Cell Fate and Function-Focus on m(6)A. Stem Cells Int. 2021;2021:8874360.
Li L, Zang L, Zhang F, Chen J, Shen H, Shu L, et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet. 2017;26:2398–411.
Cao Y, Zhuang Y, Chen J, Xu W, Shou Y, Huang X, et al. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum Mol Genet. 2020;29:727–35.
Salas IH, Burgado J, Allen NJ. Glia: victims or villains of the aging brain? Neurobiol Dis. 2020;143:105008.
Lago-Baldaia I, Fernandes VM, Ackerman SD. More Than Mortar: Glia as Architects of Nervous System Development and Disease. Front Cell Developmental Biol. 2020;8:611269.
Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng YL, Elbaz B, et al. m(6)A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron. 2020;105:293–309.e295.
Zhang T, Chen P, Li W, Sha S, Wang Y, Yuan Z, et al. Cognitive deficits in mice lacking Nsun5, a cytosine-5 RNA methyltransferase, with impairment of oligodendrocyte precursor cells. Glia. 2019;67:688–702.
Yuan Z, Chen P, Zhang T, Shen B, Chen L. Agenesis and Hypomyelination of Corpus Callosum in Mice Lacking Nsun5, an RNA Methyltransferase. Cells. 2019;8:552.
Xin Y, He Q, Liang H, et al. m6A epitranscriptomic modification regulates neural progenitor-to-glial cell transition in the retina. Elife. 2022;11:e79994.
Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, et al. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology. 2017;42:1502–10.
Merkurjev D, Hong W-T, Iida K. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21:1004–14.
Flamand MN, Meyer KD. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. 2022;50:4464–83.
Madugalle SU, Liau WS, Zhao Q, et al. Synapse-Enriched m6A-Modified Malat1 Interacts with the Novel m6A Reader, DPYSL2, and Is Required for Fear-Extinction Memory. J Neurosci. 2023;43:7084–100.
Zhang Z, Wang M, Xie D, Huang Z, Zhang L, Yang Y, et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018;28:1050–61.
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–53.
Nagayoshi Y, Chujo T, Hirata S, et al. Loss of Ftsj1 perturbs codon-specific translation efficiency in the brain and is associated with X-linked intellectual disability. Sci Adv. 2021;7:eabf3072.
Goldeck M, Gopal A, Jantsch MF, Mansouri Khosravi HR, Rajendra V, Vesely C. How RNA editing keeps an I on physiology. Am J Physiol-Cell Physiol. 2022;323:C1496–C1511.
Gowda NKC, Nawalpuri B, Ramakrishna S, Jhaveri V, Muddashetty RS. NMDAR mediated dynamic changes in m6A inversely correlates with neuronal translation. Sci Rep. 2022; 12.
Xu X, Johnson Z, Xie H. Neuronal Depolarization Induced RNA m5C Methylation Changes in Mouse Cortical Neurons. Biology. 2022;11:988.
Blaze J, Navickas A, Phillips HL, Heissel S, Plaza-Jennings A, Miglani S, et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior. Nat Commun. 2021;12.
Brande-Eilat N, Golumbic YN, Zaidan H, Gaisler-Salomon I. Acquisition of conditioned fear is followed by region-specific changes in RNA editing of glutamate receptors. Stress. 2015;18:309–18.
Zhai J, Navakkode S, Yeow SQZ, Krishna-K K, Liang MC, Koh JH, et al. Loss of CaV1.3 RNA editing enhances mouse hippocampal plasticity, learning, and memory. Proc Natl Acad Sci USA. 2022;119:e2203883119.
Koranda JL, Dore L, Shi H, Patel MJ, Vaasjo LO, Rao MN, et al. Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron. 2018;99:283.
Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, et al. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J Neurosci. 2016;36:6771.
Pupak A, Singh A, Sancho-Balsells A, Alcalá-Vida R, Espina M, Giralt A, et al. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell Mol Life Sci. 2022;79:416.
Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B, et al. N6-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1–Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol Psychiatry. 2020;88:392–404.
Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron. 2018;99:389–403.e389.
Aoki M, Watanabe Y, Yoshimoto K, Tsujimura A, Yamamoto T, Kanamura N, et al. Involvement of serotonin 2C receptor RNA editing in accumbal neuropeptide Y expression and behavioural despair. Eur J Neurosci. 2016;43:1219–28.
Mombereau C, Kawahara Y, Gundersen BB, Nishikura K, Blendy JA. Functional relevance of serotonin 2C receptor mRNA editing in antidepressant- and anxiety-like behaviors. Neuropharmacology. 2010;59:468.
Kubota-Sakashita M, Iwamoto K, Bundo M, Kato T. A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia. Mol Brain. 2014;7:5.
Xue A, Huang Y, Li M, Wei Q, Bu Q. Comprehensive analysis of differential m6A RNA methylomes in the Hippocampus of Cocaine-conditioned mice. Mol Neurobiol. 2021;58:3759–68.
Hess ME, Hess S, Meyer KD, Verhagen LAW, Koch L, Brönneke HS, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042–8.
Schmidt HD, McFarland KN, Darnell SB, Huizenga MN, Sangrey GR, Cha JHJ, et al. ADAR2-dependent GluA2 editing regulates cocaine seeking. Mol psychiatry. 2015;20:1460.
Watanabe Y, Yoshimoto K, Tatebe H, Kita M, Nishikura K, Kimura M, et al. Enhancement of alcohol drinking in mice depends on alterations in RNA editing of serotonin 2C receptors. Int J Neuropsychopharmacol. 2014;17:739–51.
Shirahase T, Watanabe Y, Tsujimura A, Kwak S, Yamamoto T, Kanamura N, et al. Ethanol preference and drinking behavior are controlled by RNA editing in the nucleus accumbens. Front Behav Neurosci. 2019;12:331.
McMillan M, Gomez N, Hsieh C, Bekier M, Li X, Miguez R, et al. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Molecular Cell. 2023;83:219–36.e7.
Zhao F, Xu Y, Gao S, Qin L, Austria Q, Siedlak SL, et al. METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol Neurodegener. 2021;16:70.
Castro-Hernandez R, Berulava T, Metelova M, Epple R, Pena Centeno T, Richter J, et al. Conserved reduction of m(6)A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts. Proc Natl Acad Sci USA. 2023;120:e2204933120.
Chen X, Yu C, Guo M, Zheng X, Ali S, Huang H, et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem Neurosci. 2019;10:2355–63.
Li H, Ren Y, Mao K, Hua F, Yang Y, Wei N, et al. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem Biophys Res Commun. 2018;498:234–9.
Lv Z, Xu T, Li R, et al. Downregulation of m6A Methyltransferase in the Hippocampus of Tyrobp-/- Mice and Implications for Learning and Memory Deficits. Front Neurosci. 2022;16:739201.
Han M, Liu Z, Xu Y, et al. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front Neurosci. 2020;14:98.
Yi-Lan Weng A, Wang X, An R, Liu K, Song H, Ming. Correspondence G-l. Epitranscriptomic m 6 A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018;97:313–25.e6.
Li Y, Guo X, Sun L, et al. N6-Methyladenosine Demethylase FTO Contributes to Neuropathic Pain by Stabilizing G9a Expression in Primary Sensory Neurons. Adv Sci. 2020;7:1902402.
Wang Y, Mao J, Wang X, Lin Y, Hou G, Zhu J, et al. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics. 2019;11:805–19.
Yu J, Zhang Y, Ma H, et al. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol Brain. 2020;13:11.
Chokkalla AK, Mehta SL, Kim TH, Chelluboina B, Kim J, Vemuganti R. Transient Focal Ischemia Significantly Alters the m6A Epitranscriptomic Tagging of RNAs in the Brain. Stroke. 2019;50:2912–21.
Xu K, Mo Y, Li D, et al. N6-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther Adv Chronic Dis. 2020;11:2040622320916024.
Zhang Z, Wang Q, Zhao X, Shao L, Liu G, Zheng X, et al. YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 2020;11:977.
Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–31.
Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–98.
Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–33.
Wright A, Vissel B. The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci. 2012;5:34.
Caglayan AO, Tuysuz B, Coskun S, Quon J, Harmanci AS, Baranoski JF, et al. A patient with a novel homozygous missense mutation in FTO and concomitant nonsense mutation in CETP. J Hum Genet. 2016;61:395–403.
Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039.
Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362:239–52.
Freude K, Hoffmann K, Jensen LR, Delatycki MB, des Portes V, Moser B, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:305–9.
de Brouwer APM, Abou Jamra R, Kortel N, Soyris C, Polla DL, Safra M, et al. Variants in PUS7 Cause Intellectual Disability with Speech Delay, Microcephaly, Short Stature, and Aggressive Behavior. Am J Hum Genet. 2018;103:1045–52.
Shaheen R, Tasak M, Maddirevula S, Abdel-Salam GMH, Sayed ISM, Alazami AM, et al. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum Genet. 2019;138:231–9.
Igoillo-Esteve M, Genin A, Lambert N, Desir J, Pirson I, Abdulkarim B, et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013;9:e1003888.
Cohen JS, Srivastava S, Farwell KD, Lu HM, Zeng W, Lu H, et al. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am J Med Genet A. 2015;167:1391–5.
Hayhurst H, de Coo IFM, Piekutowska-Abramczuk D, Alston CL, Sharma S, Thompson K, et al. Leigh syndrome caused by mutations in MTFMT is associated with a better prognosis. Ann Clin Transl Neurol. 2019;6:515–24.
Neeve VC, Pyle A, Boczonadi V, Gomez-Duran A, Griffin H, Santibanez-Koref M, et al. Clinical and functional characterisation of the combined respiratory chain defect in two sisters due to autosomal recessive mutations in MTFMT. Mitochondrion. 2013;13:743–8.
Michaud J, Kudoh J, Berry A, Bonne-Tamir B, Lalioti MD, Rossier C, et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics. 2000;68:71–79.
Braun DA, Shril S, Sinha A, Schneider R, Tan W, Ashraf S, et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am J Med Genet A. 2018;176:2460–5.
Shaheen R, Abdel-Salam GM, Guy MP, Alomar R, Abdel-Hamid MS, Afifi HH, et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015;16:210.
Blaesius K, Abbasi AA, Tahir TH, Tietze A, Picker-Minh S, Ali G, et al. Mutations in the tRNA methyltransferase 1 gene TRMT1 cause congenital microcephaly, isolated inferior vermian hypoplasia and cystic leukomalacia in addition to intellectual disability. Am J Med Genet A. 2018;176:2517–21.
Salehi Chaleshtori AR, Miyake N, Ahmadvand M, Bashti O, Matsumoto N, Noruzinia M. A novel 8-bp duplication in ADAT3 causes mild intellectual disability. Hum Genome Var. 2018;5:7.
Thomas E, Lewis AM, Yang Y, Chanprasert S, Potocki L, Scott DA. Novel Missense Variants in ADAT3 as a Cause of Syndromic Intellectual Disability. J Pediatr Genet. 2019;8:244–51.
Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49:380–5.
Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:847–55.
Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–63.
Yang J, Xu J, Zhang L, Li Y, Chen M. Identifying key m(6)A-methylated lncRNAs and genes associated with neural tube defects via integrative MeRIP and RNA sequencing analyses. Front Genet. 2022;13:974357.
Zhang L, Cao R, Li D, Sun Y, Zhang J, Wang X, et al. Ethionine-mediated reduction of S-adenosylmethionine is responsible for the neural tube defects in the developing mouse embryo-mediated m6A modification and is involved in neural tube defects via modulating Wnt/beta-catenin signaling pathway. Epigenetics Chromatin. 2021;14:52.
Gupta T, Malkin MG, Huang S. tRNA function and dysregulation in cancer. Front Cell Dev Biol. 2022;10:886642.
Song P, Tayier S, Cai Z, Jia G. RNA methylation in mammalian development and cancer. Cell Biol Toxicol. 2021;37:811–31.
Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee US. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac J Cancer Prev. 2017;18:3–9.
Cui Q, Yin K, Zhang X, Ye P, Chen X, Chao J, et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat Cancer. 2021;2:932–49.
Miao FA, Chu K, Chen HR, Zhang M, Shi PC, Bai J, et al. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Invest N. Drugs. 2019;37:1177–86.
Wang P, Wu M, Tu Z, Tao C, Hu Q, Li K, et al. Identification of RNA: 5-Methylcytosine Methyltransferases-Related Signature for Predicting Prognosis in Glioma. Front Oncol. 2020;10:1119.
Janin M, Ortiz-Barahona V, de Moura MC, Martinez-Cardus A, Llinas-Arias P, Soler M, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–74.
Huang QR, Li JW, Pan XB. A novel risk signature with 6 RNA binding proteins for prognosis prediction in patients with glioblastoma. Med (Baltim). 2021;100:e28065.
Wang B, Niu L, Wang Z, Zhao Z. RNA m1A Methyltransferase TRMT6 Predicts Poorer Prognosis and Promotes Malignant Behavior in Glioma. Front Mol Biosci. 2021;8:692130.
Huang Y, Ma J, Yang C, Wei P, Yang M, Han H, et al. METTL1 promotes neuroblastoma development through m(7)G tRNA modification and selective oncogenic gene translation. Biomark Res. 2022;10:68.
Li L, Yang Y, Wang Z, Xu C, Huang J, Li G. Prognostic role of METTL1 in glioma. Cancer Cell Int. 2021;21:633.
Dome A, Dymova M, Richter V, Stepanov G. Post-Transcriptional Modifications of RNA as Regulators of Apoptosis in Glioblastoma. Int J Mol Sci. 2022;23:9272.
Zhang S, Zhao S, Qi Y, Li B, Wang H, Pan Z, et al. SPI1-induced downregulation of FTO promotes GBM progression by regulating pri-miR-10a processing in an m6A-dependent manner. Mol Ther Nucleic Acids. 2022;27:699–717.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606 e596.
Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y. WTAP expression predicts poor prognosis in malignant glioma patients. J Mol Neurosci. 2016;60:131–6.
Pan T, Wu F, Li L, Wu S, Zhou F, Zhang P, et al. The role m(6)A RNA methylation is CNS development and glioma pathogenesis. Mol Brain. 2021;14:119.
Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019;79:5785–98.
Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021;11:480–99.
Zhu X, Yang H, Zhang M, Wu X, Jiang L, Liu X, et al. YTHDC1-mediated VPS25 regulates cell cycle by targeting JAK-STAT signaling in human glioma cells. Cancer Cell Int. 2021;21:645.
Xu C, Yuan B, He T, Ding B, Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J Cell Mol Med. 2020;24:7538–49.
Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z, et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem. 2011;286:25882–90.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90–105 e123.
Miao YQ, Chen W, Zhou J, Shen Q, Sun Y, Li T, et al. N(6)-adenosine-methyltransferase-14 promotes glioma tumorigenesis by repressing argininosuccinate synthase 1 expression in an m6A-dependent manner. Bioengineered. 2022;13:1858–71.
Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM. m(6)A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem Biol. 2021;16:324–33.
Jin DI, Lee SW, Han ME, Kim HJ, Seo SA, Hur GY, et al. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103:2102–9.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622–34.
Rocchi L, Barbosa AJ, Onofrillo C, Del Rio A, Montanaro L. Inhibition of human dyskerin as a new approach to target ribosome biogenesis. PLoS One. 2014;9:e101971.
Micaelli M, Dalle Vedove A, Cerofolini L, Vigna J, Sighel D, Zaccara S, et al. Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N (6)-Methyladenosine-Modified RNAs. ACS Pharm Transl Sci. 2022;5:872–91.
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597–601.
Moroz-Omori EV, Huang D, Kumar Bedi R, Cheriyamkunnel SJ, Bochenkova E, Dolbois A, et al. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedChem. 2021;16:3035–43.
Rivera M, Locke AE, Corre T, Czamara D, Wolf C, Ching-Lopez A, et al. Interaction between the FTO gene, body mass index and depression: meta-analysis of 13701 individuals. Br J Psychiatry. 2017;211:70.
Du T, Rao S, Wu L, Ye N, Liu Z, Hu H, et al. An association study of the m6A genes with major depressive disorder in Chinese Han population. J Affect Disord. 2015;183:279–86.
Liu Y, Zhang H. RNA m6A Modification Changes in Postmortem Nucleus Accumbens of Subjects with Alcohol Use Disorder: A Pilot Study. Genes. 2022;13:958.
Bohnsack JP, Teppen T, Kyzar EJ, Dzitoyeva S, Pandey SC. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl Psychiatry. 2019;9:34.
Karanović J, Šviković S, Pantović M, Durica S, Brajušković G, Damjanović A, et al. Joint effect of ADARB1 gene, HTR2C gene and stressful life events on suicide attempt risk in patients with major psychiatric disorders. World J Biol Psychiatry. 2015;16:261–71.
Chimienti F, Cavarec L, Vincent L, et al. Correction: Brain region-specific alterations of RNA editing in PDE8A mRNA in suicide decedents. Transl Psychiatry. 2019;9:112.
Weissmann D, van der Laan S, Underwood MD, Salvetat N, Cavarec L, Vincent L, et al. Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Transl Psychiatry. 2016;6:e878.
Breen MS, Dobbyn A, Li Q, et al. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat Neurosci. 2019;22:1402–12.
Choudhury M, Fu T, Amoah K, Jun HI, Chan TW, Park S, et al. Widespread RNA hypoediting in schizophrenia and its relevance to mitochondrial function. Sci Adv. 2023;9:eade9997.
Ye F, Wang T, Wu X, Liang J, Li J, Sheng W. N6-Methyladenosine RNA modification in cerebrospinal fluid as a novel potential diagnostic biomarker for progressive multiple sclerosis. J Transl Med. 2021;19:316.
Keller L, Xu W, Wang HX, Winblad B, Fratiglioni L, Graff C. The Obesity Related Gene, FTO, Interacts with APOE, and is Associated with Alzheimer’s Disease Risk: A Prospective Cohort Study. J Alzheimer’s Dis. 2011;23:461–9.
Khermesh K, D’Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, et al. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA. 2016;22:290.
Ma Y, Dammer EB, Felsky D, Duong DM, Klein HU, White CC, et al. Atlas of RNA editing events affecting protein expression in aged and Alzheimer’s disease human brain tissue. Nat Commun. 2021;12:1–16.
Aizawa H, Sawada J, Hideyama T, Yamashita T, Katayama T, Hasebe N, et al. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120:75–84.
Salvetat N, Checa-Robles FJ, Patel V, Cayzac C, Dubuc B, Chimienti F, et al. A game changer for bipolar disorder diagnosis using RNA editing-based biomarkers. Transl Psychiatry. 2022;12:1–10.
Lo N, Xu X, Soares F, He HH. The Basis and Promise of Programmable RNA Editing and Modification. Front Genet. 2022;13:834413.
Song J, Dong L, Sun H, Luo N, Huang Q, Li K, et al. CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons. Mol Cell. 2023;83:139–55.e139.
Adachi H, Pan Y, He X, Chen JL, Klein B, Platenburg G, et al. Targeted pseudouridylation: an approach for suppressing nonsense mutations in disease genes. Mol Cell. 2023;83:637–51.e639.
Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275:4251–7.
Kirino Y, Yasukawa T, Marjavaara SK, Jacobs HT, Holt IJ, Watanabe K, et al. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum Mol Genet. 2006;15:897–904.
Rohner E, Yang R, Foo KS, Goedel A, Chien KR. Unlocking the promise of mRNA therapeutics. Nat Biotechnol. 2022;40:1586–1600.
Anthony K. RNA-based therapeutics for neurological diseases. RNA Biol. 2022;19:176–90.
Mantile F, Prisco A. Vaccination against β-Amyloid as a Strategy for the Prevention of Alzheimer’s Disease. Biology. 2020;9:425.
Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–90.
Bohmwald K, Andrade CA, Galvez NMS, Mora VP, Munoz JT, Kalergis AM. The causes and long-term consequences of viral encephalitis. Front Cell Neurosci. 2021;15:755875.
Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. 2022;40:840–54.
Rao Y, Du S, Yang B, Wang Y, Li Y, Li R, et al. NeuroD1 induces microglial apoptosis and cannot induce microglia-to-neuron cross-lineage reprogramming. Neuron. 2021;109:4094–108.e4095.
Wang LL, Serrano C, Zhong X, Ma S, Zou Y, Zhang CL. Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell. 2021;184:5465–81 e5416.
Martier R, Konstantinova P. Gene therapy for neurodegenerative diseases: slowing down the ticking clock. Front Neurosci. 2020;14:580179.
Parambi DGT, Alharbi KS, Kumar R, Harilal S, Batiha GE, Cruz-Martins N, et al. Gene therapy approach with an emphasis on growth factors: theoretical and clinical outcomes in neurodegenerative diseases. Mol Neurobiol. 2022;59:191–233.
Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. 2022;21:52.
Lan L, Sun YJ, Jin XY, Xie LJ, Liu L, Cheng L. A Light-Controllable Chemical Modulation of m(6) A RNA Methylation. Angew Chem Int Ed Engl. 2021;60:18116–121.
Tang T, Han Y, Wang Y, Huang H, Qian P. Programmable System of Cas13-Mediated RNA Modification and Its Biological and Biomedical Applications. Front cell Dev Biol. 2021;9:677587.
Acknowledgements
We thank the members of Song and Ming laboratories for comments and suggestions.
Funding
The research in the authors’ laboratories was supported by grants from the National Institutes of Health (R35NS116843 and RF1AG079557 to HS and R35NS097370 to G-lM), Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to G-lM), and a pilot award from the Institute for RNA Innovation of the Perelman School of Medicine at the University of Pennsylvania (to HS).
Author information
Authors and Affiliations
Contributions
FZ and VI wrote the manuscript with contributions from all co-authors. GM and HS developed the outline of this paper and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, F., Ignatova, V.V., Ming, Gl. et al. Advances in brain epitranscriptomics research and translational opportunities. Mol Psychiatry (2023). https://doi.org/10.1038/s41380-023-02339-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-023-02339-x