Abstract
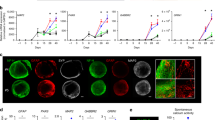
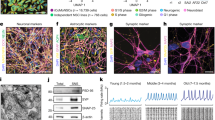
Several iPSC-derived three-dimensional (3D) cultures have been generated to model Alzheimer’s disease (AD). While some AD-related phenotypes have been identified across these cultures, none of them could recapitulate multiple AD-related hallmarks in one model. To date, the transcriptomic features of these 3D models have not been compared with those of human AD brains. However, these data are crucial to understanding the pertinency of these models for studying AD-related pathomechanisms over time. We developed a 3D bioengineered model of iPSC-derived neural tissue that combines a porous scaffold composed of silk fibroin protein with an intercalated collagen hydrogel to support the growth of neurons and glial cells into complex and functional networks for an extended time, a fundamental requisite for aging studies. Cultures were generated from iPSC lines obtained from two subjects carrying the familial AD (FAD) APP London mutation, two well-studied control lines, and an isogenic control. Cultures were analyzed at 2 and 4.5 months. At both time points, an elevated Aβ42/40 ratio was detected in conditioned media from FAD cultures. However, extracellular Aβ42 deposition and enhanced neuronal excitability were observed in FAD culture only at 4.5 months, suggesting that extracellular Aβ deposition may trigger enhanced network activity. Remarkably, neuronal hyperexcitability has been described in AD patients early in the disease. Transcriptomic analysis revealed the deregulation of multiple gene sets in FAD samples. Such alterations were strikingly similar to those observed in human AD brains. These data provide evidence that our patient-derived FAD model develops time-dependent AD-related phenotypes and establishes a temporal relation among them. Furthermore, FAD iPSC-derived cultures recapitulate transcriptomic features of AD patients. Thus, our bioengineered neural tissue represents a unique tool to model AD in vitro over time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Datasets generated and analyzed in this study can be found in the published article and its supplementary information files. Supplementary information is available at MP’s website. Additional information are available from the corresponding author. ROSMAP resources can be requested at https://www.radc.rush.edu. The NanoString data has been stored on Synapse and is available at https://doi.org/10.7303/syn51471664.
Material availability
A materials transfer agreement covers the TG3 antibody provided by the Feinstein Institutes for Medical Research and the Albert Einstein College of Medicine.
References
Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1:a006189.
Qiu C, Kivipelto M, von Strauss E. Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci. 2009;11:111–28.
D’Avanzo C, Aronson J, Kim YH, Choi SH, Tanzi RE, Kim DY. Alzheimer’s in 3D culture: challenges and perspectives. Bioessays. 2015;37:1139–48.
Penney J, Ralvenius WT, Tsai LH. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol Psychiatry. 2020;25:148–67.
Cenini G, Hebisch M, Iefremova V, Flitsch LJ, Breitkreuz Y, Tanzi RE, et al. Dissecting Alzheimer’s disease pathogenesis in human 2D and 3D models. Mol Cell Neurosci. 2021;110:103568.
Lagomarsino VN, Pearse RV, Liu L, Hsieh Y-C, Fernandez MA, Vinton EA, et al. Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron. 2021;109:3402–3420.e3409.
Cantley W, Du C, Lomoio S, DePalma T, Peirent E, Kleinknecht D, et al. Functional and sustainable 3D human neural network models from pluripotent stem cells. ACS Biomater Sci Eng. 2018. https://doi.org/10.1021/acsbiomaterials.8b00622.
Rouleau N, Cantley WL, Liaudanskaya V, Berk A, Du C, Rusk W, et al. A long-living bioengineered neural tissue platform to study neurodegeneration. Macromol Biosci. 2020;20:e2000004.
Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–36.
Hector A, Brouillette J. Hyperactivity induced by soluble amyloid-beta oligomers in the early stages of Alzheimer’s disease. Front Mol Neurosci. 2020;13:600084.
Wan YW, Al-Ouran R, Mangleburg CG, Perumal TM, Lee TV, Allison K, et al. Meta-analysis of the Alzheimer’s disease human brain transcriptome and functional dissection in mouse models. Cell Rep. 2020;32:107908.
Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimers Dis. 2018;64:S161–S189.
Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc. 2011;6:1612–31.
Shankar GM, Welzel AT, McDonald JM, Selkoe DJ, Walsh DM. Isolation of low-n amyloid beta-protein oligomers from cultured cells, CSF, and brain. Methods Mol Biol. 2011;670:33–44.
Du C, Collins W, Cantley W, Sood D, Kaplan DL. Tutorials for electrophysiological recordings in neuronal tissue engineering. ACS Biomater Sci Eng. 2017;3:2235–46.
Iovino M, Patani R, Watts C, Chandran S, Spillantini MG. Human stem cell-derived neurons: a system to study human tau function and dysfunction. PLoS ONE. 2010;5:e13947.
Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data. 2016;3:160089.
Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q, et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data. 2018;5:180185.
Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci. 2018;21:811–9.
Preuss C, Pandey R, Piazza E, Fine A, Uyar A, Perumal T, et al. A novel systems biology approach to evaluate mouse models of late-onset Alzheimer’s disease. Mol Neurodegener. 2020;15:67.
Pandey RS, Graham L, Uyar A, Preuss C, Howell GR, Carter GW. Genetic perturbations of disease risk genes in mice capture transcriptomic signatures of late-onset Alzheimer’s disease. Mol Neurodegener. 2019;14:50.
Muratore CR, Srikanth P, Callahan DG, Young-Pearse TL. Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS ONE. 2014;9:e105807.
Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88–95.
Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, et al. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS ONE. 2010;5:e11853.
Muratore CR, Zhou C, Liao M, Fernandez MA, Taylor WM, Lagomarsino VN, et al. Cell-type dependent Alzheimer’s disease phenotypes: probing the biology of selective neuronal vulnerability. Stem Cell Rep. 2017;9:1868–84.
Engle SJ, Blaha L, Kleiman RJ. Best practices for translational disease modeling using human iPSC-derived neurons. Neuron. 2018;100:783–97.
Pettinato G, Wen X, Zhang N. Formation of well-defined embryoid bodies from dissociated human induced pluripotent stem cells using microfabricated cell-repellent microwell arrays. Sci Rep. 2014;4:7402.
Topol A, Tran NN, Brennand KJ. A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. J Vis Exp. 2015;96:e52495.
Readhead B, Hartley BJ, Eastwood BJ, Collier DA, Evans D, Farias R, et al. Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nat Commun. 2018;9:4412.
Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18.
Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–98.
Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–23.
Liao M-C, Muratore CR, Gierahn TM, Sullivan SE, Srikanth P, De Jager PL, et al. Single-cell detection of secreted Aβ and sAPPα from human IPSC-derived neurons and astrocytes. J Neurosci. 2016;36:1730–46.
Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–5.
Arber C, Toombs J, Lovejoy C, Ryan NS, Paterson RW, Willumsen N, et al. Familial Alzheimer’s disease patient-derived neurons reveal distinct mutation-specific effects on amyloid beta. Mol Psychiatry. 2020;25:2919–31.
O’Connor A, Pannee J, Poole T, Arber C, Portelius E, Swift IJ, et al. Plasma amyloid-beta ratios in autosomal dominant Alzheimer’s disease: the influence of genotype. Brain. 2021;144:2964–70.
Eimer WA, Vassar R. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Abeta42 accumulation and Caspase-3 activation. Mol Neurodegener. 2013;8:2.
Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–26.
Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–103.
Iovino M, Agathou S, Gonzalez-Rueda A, Del Castillo Velasco-Herrera M, Borroni B, Alberici A, et al. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain. 2015;138:3345–59.
Jicha GA, Lane E, Vincent I, Otvos L Jr, Hoffmann R, Davies P. A conformation- and phosphorylation-dependent antibody recognizing the paired helical filaments of Alzheimer’s disease. J Neurochem. 1997;69:2087–95.
Kittelberger KA, Piazza F, Tesco G, Reijmers LG. Natural amyloid-beta oligomers acutely impair the formation of a contextual fear memory in mice. PLoS ONE. 2012;7:e29940.
Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167.
Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53.
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125.
Freidin M, Bennett MV, Kessler JA. Cultured sympathetic neurons synthesize and release the cytokine interleukin 1 beta. Proc Natl Acad Sci USA. 1992;89:10440–3.
Sebire G, Emilie D, Wallon C, Hery C, Devergne O, Delfraissy JF, et al. In vitro production of IL-6, IL-1 beta, and tumor necrosis factor-alpha by human embryonic microglial and neural cells. J Immunol. 1993;150:1517–23.
Lim JC, Lu W, Beckel JM, Mitchell CH. Neuronal release of cytokine IL-3 triggered by mechanosensitive autostimulation of the P2X7 receptor is neuroprotective. Front Cell Neurosci. 2016;10:270.
De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci Data. 2018;5:180142.
Gonzalez C, Armijo E, Bravo-Alegria J, Becerra-Calixto A, Mays CE, Soto C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry. 2018;23:2363–74.
Mertens J, Reid D, Lau S, Kim Y, Gage FH. Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu Rev Genet. 2018;52:271–93.
Golde TE. Alzheimer’s disease - the journey of a healthy brain into organ failure. Mol Neurodegener. 2022;17:18.
Ghatak S, Dolatabadi N, Trudler D, Zhang X, Wu Y, Mohata M, et al. Mechanisms of hyperexcitability in Alzheimer’s disease hiPSC-derived neurons and cerebral organoids vs isogenic controls. Elife. 2019;8:e50333.
Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, et al. Bioengineered functional brain-like cortical tissue. Proc Natl Acad Sci USA. 2014;111:13811–6.
Hronik-Tupaj M, Raja WK, Tang-Schomer M, Omenetto FG, Kaplan DL. Neural responses to electrical stimulation on patterned silk films. J Biomed Mater Res A. 2013;101:2559–72.
Tang-Schomer MD, Hu X, Tupaj M, Tien LW, Whalen M, Omenetto F, et al. Film-based implants for supporting neuron-electrode integrated interfaces for the brain. Adv Funct Mater. 2014;24:1938–48.
Ramos DM, Skarnes WC, Singleton AB, Cookson MR, Ward ME. Tackling neurodegenerative diseases with genomic engineering: a new stem cell initiative from the NIH. Neuron. 2021;109:1080–3.
Acknowledgements
We thank Dr. D. Selkoe (Brigham and Women’s Hospital, Boston) for kindly providing us with the R1282 antibody. We also thank Rachel Willen, Edward K. Robinson, Griffin Sigal, and Isabel Paine for their technical support.
Funding
This work was supported by awards from the National Institutes of Health: 1R21AG065792 (to GT), 5R01AG061838 (to GT, PGH, and DLK), R01AG055909 (to TLYP), and U54AG054345 (to GWC). ROSMAP is supported by P30AG10161, P30AG72975, R01AG15819, R01AG17917. U01AG46152, U01AG61356 (to DAB).
Author information
Authors and Affiliations
Contributions
The authors confirm contributions to the manuscript as follows: conceptualization: SL, GT; data curation: SL, RSP, NR; formal analysis: SL, RSP, NR; investigation: SL, NR, BM, WK, WLC; methodology: SL, RSP, NR, GWC; resources: WLC, GWC, DAB, TLYP, DLK; project administration: GT; supervision: SL, GWC, DLK, GT; validation: SL, RSP, NR, BM, WK, GT; visualization: SL, RSP, NR; writing - original draft preparation: SL, RSP, NR, GT; writing - review and editing: SL, RSP, NR, BM, WK, WLC, PGH, DAB, TLYP, GWC, DLK, GT.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lomoio, S., Pandey, R.S., Rouleau, N. et al. 3D bioengineered neural tissue generated from patient-derived iPSCs mimics time-dependent phenotypes and transcriptional features of Alzheimer’s disease. Mol Psychiatry 28, 5390–5401 (2023). https://doi.org/10.1038/s41380-023-02147-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02147-3
This article is cited by
-
Modeling the neuroimmune system in Alzheimer’s and Parkinson’s diseases
Journal of Neuroinflammation (2024)