Abstract
Lithium (Li) is recommended for long-term treatment of bipolar disorder (BD). However, its mechanism of action is still poorly understood. Induced pluripotent stem cell (iPSC)-derived brain organoids have emerged as a powerful tool for modeling BD-related disease mechanisms. We studied the effects of 1 mM Li treatment for 1 month in iPSC-derived human cortical spheroids (hCS) from 10 healthy controls (CTRL) and 11 BD patients (6 Li-responders, Li-R, and 5 Li non-treated, Li-N). At day 180 of differentiation, BD hCS showed smaller size, reduced proportion of neurons, decreased neuronal excitability and reduced neural network activity compared to CTRL hCS. Li rescued excitability of BD hCS neurons by exerting an opposite effect in the two diagnostic groups, increasing excitability in BD hCS and decreasing it in CTRL hCS. We identified 132 Li-associated differentially expressed genes (DEGs), which were overrepresented in sodium ion homeostasis and kidney-related pathways. Moreover, Li regulated secretion of pro-inflammatory cytokines and increased mitochondrial reserve capacity in BD hCS. Through long-term Li treatment of a human 3D brain model, this study partly elucidates the functional and transcriptional mechanisms underlying the clinical effects of Li, such as rescue of neuronal excitability and neuroprotection. Our results also underscore the substantial influence of treatment duration in Li studies. Lastly, this study illustrates the potential of patient iPSC-derived 3D brain models for precision medicine in psychiatry.
Similar content being viewed by others
Introduction
Bipolar disorder (BD) is characterized by recurrent episodes of mania and depression and has a prevalence of 1–2% in the general population [1]. The lack of an accurate in vitro model remained a challenge for BD research, but the introduction of induced pluripotent stem cells (iPSCs) [2] has provided a powerful technique for modeling disease etiology and treatment. In particular, patient-derived iPSCs have been demonstrated to be a feasible model for studying in vitro the transcriptional and functional effects of several mood stabilizers commonly prescribed to treat BD [3,4,5].
Lithium (Li) is considered the treatment of choice for long-term relapse prevention, having a full response rate of ~30% [6], but with a poorly understood mechanism of action. Li has been found to selectively reverse the hyperexcitable phenotype of iPSC-derived neurons from Li-responders (Li-R) [3], and to increase mitochondrial respiration in Li-R patient iPSC-derived neural precursor cells (NPCs) [5]. Additionally, the transcriptional effects of Li have been studied in multiple cell types like peripheral blood mononuclear cells [7], and iPSC-derived neurons [3, 4] and NPCs [5]. Nevertheless, to our knowledge, Li transcriptional effects have not yet been addressed in complex 3-dimensional (3D) cell cultures such as brain organoids derived from BD iPSCs. In recent years, brain organoids have emerged as a promising tool for neurodevelopmental and brain disease modeling in vitro [8,9,10]. Indeed, 3D cell cultures are regarded as biologically more relevant than 2-dimensional (2D) cell cultures, as they partly recreate in vitro the complex architecture and diverse cell type composition found in vivo [8], allowing for a more informative analysis of the biological processes related to illness and therapeutic interventions.
Since brain cortex abnormalities are strongly associated with BD [11], we specifically generated iPSC-derived human cortical spheroids (hCS) from healthy controls (CTRL) and BD patients, and studied the effects of Li treatment at the transcriptional level by RNA-sequencing analysis and at the functional level by patch-clamp electrophysiology, calcium imaging, mitochondrial assays and cytokine profiling. We hypothesized that Li would differently impact the biochemistry and cellular neurophysiology of CTRL and BD hCS.
Methods
Donor samples
Donors were recruited through the Norwegian TOP (Thematically Organized Psychosis) study. Recruitment procedures, inclusion and exclusion criteria and clinical assessments for the TOP study have been described elsewhere [12, 13]. Fibroblasts from skin biopsies were isolated from 10 CTRL and 11 BD patients selected based on clinical information. Patients experiencing no symptoms or only mild symptoms during Li treatment (BD#4–6 and #9–11) were considered Li responders (Li-R), while the other patients (BD#1–3 and #7–8), whose responsiveness to Li was either not confirmed or not known, were considered non-treated (Li-N) (Supplementary Table 1). Li-R group included a partial responder (BD#4), due to periods of poor Li compliance. Details of several donors used in this study (CTRL#1–3 and BD#1–6) have previously been described [5]. Groups were gender-balanced and average age was not significantly different. Supplementary Table 1 summarizes the clinical characteristics of the study participants. All participants have given written consent and the study was approved by the Norwegian Data Protection Agency and the Regional Ethics Committee of the South-Eastern Norway Regional Health Authority (REK grant: #2012/2204). The authors declare that all procedures contributing to this work comply with the ethical standards of relevant guidelines and regulations.
IPSC derivation and cortical spheroids generation
Fibroblast from all donors were grown, expanded and reprogrammed as previously described [5]. Each iPSC line was subjected to rigorous quality control by phenotyping, regular monitoring of morphology and pluripotency marker expressions. KaryoStat GeneChip array (ThermoFisher) was used for karyotyping of iPSCs at passages 15–20 for digital visualization of chromosome aberrations [5]. All iPSCs were differentiated to cortical spheroids (hCS) following a previously published protocol [8], with proved high reproducibility. Pictures were taken at different time points for determining organoids size progression and Fiji software was used for area counting. Images were converted to 8-bits and binary color, scale was set using as reference distance 51.8 mm of diameter of the culture dish, particles were analyzed counting the area, and volume in mm³ was estimated using the formula: \(V = \frac{4}{3}\pi ( {\surd \frac{{{{\rm{area}}}}}{\pi }} )^3\).
Lithium administration
The hCS from all 21 donors were treated with the clinically recommended therapeutic concentration of 1 mM lithium chloride (Li, Sigma L4408), diluted in sterile Milli-Q water, which was used as vehicle control. Li brain concentration is usually moderately lower than the serum concentration, with an estimated mean brain/serum ratio of 0.78 ± 0.26 [14]. However, we used 1 mM Li for the experiments in accordance with previous in vitro studies [3, 5, 15]. This level is within the therapeutic range (approx. 0.6–1.2 mM) of serum concentrations that are usually targeted during Li treatment of BD patients. Treatment duration was 1 month, from day 150 until day 180, with medium and drug refreshment every 3–4 days. We treated all hCS with Li, regardless of their diagnosis or prescribed treatments, to identify genes and underlying mechanisms associated with Li independent of diagnosis.
RT-PCR and TaqMan-array cards
At day 180, RNA samples were collected from hCS and cells were lysed with lysis buffer. Total RNA was extracted with the RNeasy Plus Mini kit (Qiagen, #74136). Reverse transcription and RT-PCR and mRNA expression calculation were performed as previously described [5]. Primers used for iPSCs characterization are specified in Supplementary Table 2. For day 180 hCS characterization, it was used a Custom Gene Expression TaqMan Array Card - Format 16 (AppliedBiosystems, #4346798), with primers for detection of the genes in Supplementary Table 3.
Cryosectioning and immunohistochemistry
CTRL and BD hCS were fixed at day 180 in 4% paraformaldehyde in PBS O/N at 4 °C, transferred to 30% sucrose for 24 h, embedded into optimum cutting temperature (OCT) compound (Sakura Finetek, #4583) and stored at −80 °C. Each line was cut in sections 10 µm thick with a cryostat (Leica) and stainings were performed as previously described [5]. Antibodies and dilutions are specified in Supplementary Table 4. Images were acquired using a Zeiss-LSM700 confocal microscope and processed using Fiji software. At least 300 cells were analyzed for each marker from all lines.
Electrophysiology
For whole-cell patch-clamp experiments, CTRL and BD hCS were sliced (150 μm thickness) using a vibrating microtome (VT1000S, Leica Biosystems, USA), and slices were kept at room temperature in HEPES-based ACSF composed of 135 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM HEPES, 10 mM d-glucose; pH 7.35; 300–310 mOsm/L. Whole-cell patch-clamp electrophysiology recordings (whole cell currents, potentials and spontaneous excitatory postsynaptic currents, sEPSCs), data evaluation and analysis were performed as previously described [16]. The Nernst equation \(V_{{{\rm{rev}}}} = \frac{{RT}}{{zF}}{{{{{\mathrm{ln}}}}}}( {\frac{{[ X ]{{{\rm{out}}}}}}{{[ X ]{{{\rm{in}}}}}}} )\) was used for the theoretical calculation of the reversal potential for Cl− (Vrev) for estimation of potential impact of inhibitory spontaneous synaptic currents to PSCs recording at holding potential −60 mV. The theoretical reversal potential for Cl− in applied experimental conditions was calculated as V rev = −75.61 mV, that does not allow the activation of inward Cl−-mediated currents under Vh = −60 mV.
Live calcium imaging
For live calcium imaging, hCS were sliced similar to patch-clamp with a vibrating microtome (150 μm thickness). Slices were allowed to recover for at least 1 h at room temperature in HEPES-based ACSF composed of 135 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM HEPES, 10 mM d-glucose pH 7.35 with an osmolarity of 310 mOsm. For Ca2+ imaging, slices were loaded with 1 μM Cal-520, AM (Abcam). Recordings, data evaluation and analysis were performed as previously described [17]. A custom-made MATLAB (Math Works, USA) code was used to analyze calcium traces (deposited in Github: https://github.com/Zluglu/CalciumSignalAnalysis.git). Two hundred and ninety-one recordings of calcium transients (102 CTRL and 189 BD) were generated and 50–200 ROIs per recording were analyzed.
RNA extraction, sequencing and data processing
Total RNA was extracted from hCS using the RNeasy Plus Mini Kit (Qiagen). Library preparation and paired-end RNA-sequencing were performed at the Norwegian High-Throughput Sequencing Centre (www.sequencing.uio.no). Briefly, libraries were prepared with Illumina TruSeq Stranded mRNA kit, and sequenced as a single batch on an Illumina NovaSeq S4 platform at an average depth of 50 million reads per sample, using a read length of 100 bp and an insert size of 350 bp. Raw sequencing reads were quality assessed with FastQC (Babraham Institute) and further processed with Trimmomatic V0.32 [18]. HISAT2 [19] was then used to map the trimmed reads to the human GRCh38 reference genome. To quantify gene expression levels, mapped reads were summarized at the gene level using featureCounts [20] guided by ENSEMBL annotations.
Cell type deconvolution and comparison with in vivo spatiotemporal brain expression
Computational estimation of cell type abundances (deconvolution) was performed with CIBERSORTx [21] using the web-interface (https://cibersortx.stanford.edu/) with default parameters, selecting 500 permutations for statistical testing. We used as reference the expression signature from ten cell types obtained from single-cell RNA sequencing of three of our 180 days old CTRL hCS using the Chromium Next GEM Single Cell 3′ v3.1 (10X Genomics). Data from the CTRLs was integrated into a merged dataset, consisting of 11,649 cells. Normalization and integration were performed with Harmony and Seurat v4 SCT. Cluster annotation was performed with GSEA together with several automatic cell type prediction packages. To assess the extent to which in vitro hCS transcriptional profiles match the gene expression profiles of human primary in vivo brain tissue, gene-level spatiotemporal RNA sequencing data sets were downloaded from the BrainSpan resource (https://www.brainspan.org/static/download.html) [22, 23]. Concordance analyses were performed using Spearman’s correlation after converting gene counts to RPKM values and filtering out non-expressed and low-abundance transcripts.
Differential expression (DE) analysis and gene ontology (GO) enrichment tests
Genes were pre-filtered using filtByExpr function in edgeR package [24], keeping genes with a count per million above k in n samples, where k is determined by the library size and n is determined by the smallest group sample size [25]. Since KaryoStat analysis revealed chromosomal aberrations in several of the iPSC lines (Supplementary Fig. 1), genes located within affected regions were excluded prior to analysis (642 genes; Supplementary Table 5). For DE analyses, the R packages DESeq2 [26] and limma-voom [27] were used. While DESeq2 typically identifies a larger number of differentially expressed genes (DEGs) than limma-voom, the latter allows for more complex design matrices. To increase the number of significant genes, any DEG identified by either of the two packages was considered significant and retained for downstream analyses. This strategy was deemed appropriate as the two methods were found to yield highly concordant results (Supplementary Fig. 6a, b). Since a paired design was used in all DE analyses comparing Li-treated hCS against non-treated hCS, demographic variables were perfectly matched across groups and only neuronal proportions were adjusted for. Otherwise, sex and age were included as covariates in addition to neuron abundance. A DEG was considered significant if the FDR was <0.10. Gene Ontology (GO) enrichment tests of significant DEG sets were conducted with clusterProfiler [28] selecting BP (biological processes) as the ontology of interest. A GO term was considered significantly enriched if the FDR was <0.10. We performed gene set enrichment analysis (GSEA) using the R package clusterProfiler [28] and enrichment analysis of synaptic functions using SynGO (https://www.syngoportal.org/).
ELISA cytokine quantification
Supernatants were collected at different time points from all 21 donor hCS (10 CTRL and 11 BD) and stored at −80 °C until analysis. Samples were not diluted before analysis. Cytokine levels in supernatants were measured with U-PLEX Biomarker Group 1 (hu) Assay (MSD, K15067L-1), customized for detection of human IL-1β, IL-6 and TNF-α. Cytokine concentration values were corrected by the estimated volume (in cm3) of hCS tissue in each plate estimated with ImageJ as indicated in the section for hCS.
Bioenergetic assessment
Cellular bioenergetics assessment of hCS was performed using a Seahorse XFe24 extracellular flux analyzer (Agilent SeahorseXF Analysers) [5]. Briefly, hCS were sliced (150 μm thickness), and allowed to recover for 5–10 minutes at room temperature in HEPES-based ACSF. Slices were transferred to unbuffered media in a XFe24-well plate coated with poly-L-lysine (Sigma, #P8920), and 5–6 pieces were plated per well. Mitochondrial respiration function was recorded as previously described [5]. All experiments were performed in technical duplicates and repeated at least twice. Values were normalized to the total protein content in each well of the plate using the BCA protein assay kit (ThermoFisher, #23225).
Statistical analysis
One batch of organoids was differentiated for each of the 21 donor hiPSCs, and two organoids per donor were used for each read-out, except for RNA-seq analysis, which was performed using one organoid per donor and condition. Data are presented as mean ± SD for organoid size and IHC, and as mean ± SEM for electrophysiology, calcium imaging, Seahorse and cytokine analysis. Statistical significance was tested using either one-way analysis of variance (ANOVA) between groups for the transcriptional analysis or ANCOVA for spatiotemporal comparisons. Mann-Whitney U test and Wilcoxon test were used for unpaired and paired group comparisons respectively for calcium imaging, whole-cell patch clamp electrophysiology, mitochondrial parameters, hCS size and IHC quantifications. As the distribution of cytokine levels was skewed, data was log10 transformed before performing the univariate regression analysis for assessing cytokine levels at different time points evaluating time, group, and time × group effects. Regression model between cytokine and mitochondrial values with estimated cellular fractions and hCS size was performed by first carrying out a partial regression adjusting for donor id, treatment and diagnosis group (i.e. patient or control) as covariates. Variables with p < 0.2 were then included in a stepwise regression, where donor id, treatment and diagnostic group were also included and adjusted for.
Results
Differentiation of CTRL and BD iPSCs to hCS
Fibroblasts from skin biopsies were obtained from 10 CTRL and 11 BD patients, including 5 Li-N and 6 Li-R donors, and reprogrammed to iPSCs. One clone from each donor was differentiated to hCS (Figs. 1 and 2a). Details about donor age, gender and diagnosis are provided in Supplementary Table 1.
Fibroblasts isolated from 10 controls (CTRL) and 11 bipolar disorder (BD) patients, including 5 lithium non-treated (Li-N) and 6 Li-responders (Li-R), were reprogrammed to induced pluripotent stem cells (iPSCs) and differentiated to cortical spheroids (hCS). All 21 hCS lines were treated with 1 mM lithium (Li) and water as vehicle control. Medium-to-long-term effects of drug exposure (1 month) were assessed through transcriptional profiling and functional analyses in 180-day-old hCS.
a Representative images of CTRL and BD hCS from both Li-N and Li-R patients at days 30, 90 and 150 of differentiation. b Growth of CTRL and BD hCS size through differentiation show decreased volume at most time points of Li-N and Li-R hCS compared to CTRL hCS. Individual hCS volumes were estimated from their areas measured with ImageJ. c Li induced growth after 1 month treatment from day 150 until day 180. Both CTRL and Li-R, but not for Li-N hCS, present a significant increase in size. d Representative immunohistochemistry images of CTRL, Li-N and Li-R hCS with and without Li treatment. Fluorescence was measured for cytoplasmic MAP2 (e) and SV2A (f), by counting signal intensity with ImageJ. Results are presented in arbitrary units (a.u.). CTRL (N = 10), Li-N (N = 5), Li-R (N = 6). Data was analyzed by Mann–Whitney U test and Wilcoxon test for unpaired and paired group comparisons respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
All 21 iPSC lines expressed pluripotency markers OCT4, SOX2, NANOG and alkaline phosphatase (AP) (Supplementary Fig. 1a–g). Karyotyping revealed a partial loss in three patient lines at the 6q24.3 locus (Supplementary Fig. 1f and Supplementary Table 5). Furthermore, another patient line harbored a 98.7 kb and a 39.6 kb gain in the 1q21.3 and 5p15.31 loci respectively, and a CTRL line a 6.97 kb loss in the 6p24.3 locus (Supplementary Fig. 1c). These chromosome gains and losses included ~642 genes with detectable expression levels in the hCS (Supplementary Table 5). Consequently, these genes were excluded before conducting the DE analyses.
Average size of the BD hCS was lower than CTRL hCS, in particular at later stages of differentiation (Fig. 2b), in line with MRI findings of thinner cortical regions in BD [11, 29]. At day 180 of differentiation, RT-PCR analysis of cortex development and maturation markers revealed decreased expression in CTIP2, SATB2, TBR1 and SYN1 in Li-N hCS compared to CTRL hCS (Supplementary Fig. 2a).
All hCS were treated with 1 mM Li for 1 month from differentiation day 150–180. This treatment duration was chosen because full therapeutic effects typically require at least 10–21 days [30]. Li treatment produced a significant increase in size in both CTRL and Li-R hCS (Fig. 2c), in line with studies showing that Li increases neurogenesis in the dentate gyrus in rodent hippocampus [31], and is associated with thicker cortex and larger subcortical volumes in BD patients [32].
Immunohistochemistry of day 180 hCS revealed no differences in the synaptic density marker synaptic vesicle protein 2A (SV2A) nor the neuron maturity marker microtubule-associated protein 2 (MAP2) between BD and CTRL (Fig. 2d–f), but Li increased MAP2 and SV2A levels in CTRL hCS and increased SV2A in Li-N hCS (Fig. 2e, f), suggesting improved neuronal maturation.
Li treatment rescues neuronal excitability in BD hCS
We performed whole-cell patch-clamp recordings of 180-day-old hCS to characterize hCS neurons and functionally assess diagnosis-specific differences and Li treatment effects in vitro (Fig. 3a–g and Supplementary Fig. 2b–h). Evoked whole-cell currents (Fig. 3a) demonstrated that BD hCS neurons were less excitable than CTRL hCS neurons, with right-shifted current–voltage (IV) curves (Fig. 3c) toward higher membrane depolarization values (threshold voltage, Vt, Fig. 3e and Supplementary Table 6) required for maximum whole-cell current (Imax) induction. Li-N and Li-R showed similar characteristics in IV curves. Li treatment had an opposite effect in CTRL and BD hCS neurons, decreasing excitability in CTRL hCS (right-shifting IV curves and Vt shift, not significant) and increasing excitability in BD hCS (left-shifting IV curves, Fig. 3c and Supplementary Fig. 2b) and decreasing Vt significantly in Li-N hCS (Fig. 3e, Supplementary Fig. 2e). Li treatment increased currents in BD hCS, mainly driven by effects in Li-R hCS (Fig. 3d, Supplementary Fig. 2d and Supplementary Table 6).
a Raw traces of whole-cell currents (holding potential Vh = −60 to −70 mV) representative for each condition. b Raw traces of action potentials (AP) representative for each condition. c Current–voltage (IV) curves for each group. Currents of individual IV curves were normalized to maximum peak values and then averaged. d Evoked whole-cell currents peak maximum values (Imax). Li significantly increases Imax in BD hCS (Δ(Imax) = 171.94 ± 76.80 pA, p = 0.01), particularly for Li-R hCS neurons (Δ(Imax) = 209.34 ± 102.42 pA, p = 0.03). e Threshold membrane voltages (Vt) of Imax, showing BD is less excitable than CTRL hCS neurons (Δ(Vt) = 5.32 ± 2.93 mV, p = 0.04). Li treatment increases excitability of BD hCS (via decreasing threshold voltage, Δ(Vt) = 5.14 ± 2.86 mV, p = 0.02), significant for Li-N hCS (Δ(Vt) = 11.84 ± 5.62 mV, p = 0.02). f Membrane resting potentials (RP). g Voltage–current (VI) curves for each group. Individual VI curves were averaged for correspondent group. h Representative trace images of spontaneous CaCl2 transients in hCS regions of interest (ROIs) over a 20-min period. i Percentage of signaling ROIs after Li treatment. j Average amplitude of Ca2+ signal transients (%ΔF/F). I-shaped box charts represents 25–75% of range intervals and median horizontal brackets, overlapped with data; square symbol represent mean value. CTRL (N = 10), Li-N (N = 5), Li-R (N = 6). Data was analyzed by Mann–Whitney U test and Wilcoxon test for unpaired and paired group comparisons respectively. *P < 0.05, **P < 0.01.
In line with voltage-clamp recordings, current-clamp recordings (Fig. 3b) revealed similar excitability differences (CTRL hCS are more excitable due to left-shifted voltage-current (VI) curves, Fig. 3g) and changes upon treatment (Li decreased excitability in CTRL hCS and increased it in BD hCS). Evoked action potentials (AP) appeared at similar amplitudes of threshold currents (It), and Li treatment did not affect It (Supplementary Fig. 2h). Despite comparable membrane resting potentials (RP) for CTRL and BD hCS (Fig. 3f and Supplementary Fig. 2f), Li significantly decreased RP only in CTRL hCS. Input resistance (Rin) was analogous between groups, and Li treatment had no significant effect.
Next, we studied Li effects on synaptic activity and connectivity by recording spontaneous excitatory postsynaptic currents (sEPSC), and performed distribution analysis for sEPSC amplitudes and frequencies (Supplementary Fig. 3a–k and Supplementary Table 7). Li-R exhibited higher sEPSC amplitudes compared to CTRL and Li-N, and sEPSC frequencies were similar between groups. Li treatment resulted in a non-significant increase of sEPSC amplitudes for CTRL and decrease for Li-R hCS, and non-significant increase of sEPSC frequencies in all groups, indicating a tendency of increased synaptic activity.
To assess whether the decreased excitability of single neurons in BD hCS (Fig. 3c, g) could modify neural network activity we measured somatic Ca2+ transient events (Fig. 3h–j and Supplementary Fig. 4a–j). In untreated hCS, BD hCS showed significant decrease in percentage of signaling cells per region of interest (ROI), driven by the Li-N group (Fig. 3i and Supplementary Fig. 4a) and the amplitude of signal events compared to CTRL hCS (Fig. 3j and Supplementary Fig. 4b), suggesting decreased neural network activity. Li treatment decreased Ca2+ signal amplitudes in all groups, but this was only significant in CTRL hCS (Fig. 3j).
Cell type deconvolution and spatiotemporal gene expression analysis of hCS
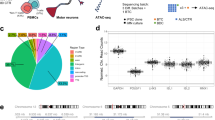
We next assessed the effect of Li on global gene expression. Principal component analysis (PCA) showed weak clustering of samples across diagnostic and response groups (Fig. 4a). Moreover, evaluation of treated and untreated pairs showed that Li treatment had mostly small effects across donors (Fig. 4b). This was confirmed by variance partition analysis, which attributed only small effects to Li responsiveness and gender (Fig. 4c). As expected, cell type composition and donor effects were the most important sources of variation in gene expression (Fig. 4c).
a Principal component analysis (PCA) showing a strong donor effects and weaker clustering across diagnostic and response categories. b The same PCA plot displaying pairing information between untreated (circles) and treated (triangles) pairs. c Variance partition plot showing the proportion of gene expression variance attributed to different sources. Residuals constitute additional, unknown sources of variation not accounted for. d Estimation of cell type fractions in untreated CTRL and BD hCS. Significant differences detected in mature neurons and interneurons proportions. Data was analyzed by Mann–Whitney U test for group comparisons. e Overlap between in vitro gene expression (hCS) and in vivo human spatial brain expression profiles (BrainSpan). Brain regions nomenclature and abbreviations is the same as in [71]. f Overlap between in vitro gene expression and in vivo human temporal brain expression profiles. Strong correspondence seen between hCS and mid fetal stages (*p < 0.05, **p < 0.01, ANCOVA). CTRL (N = 10), Li-N (N = 5), Li-R (N = 6). OPCs oligodendrocyte precursor cells, RGs radial glia cells, NPCs neural precursor cells.
Computational deconvolution of cell type proportions showed that CTRL hCS had a substantial increase in mature neurons and interneurons compared to BD hCS (Fig. 4d). Response-status showed significant effect on several cell fractions in Li-N hCS, including reduced neuronal types and increased RGs and intermediate progenitors, while Li treatment did not have a significant effect on cell type estimates (Supplementary Fig. 5a, b).
To determine the extent to which hCS gene expression profiles resemble human developmental brain profiles in vivo, spatiotemporal comparisons were performed using the BrainSpan dataset [22, 23]. Both CTRL and BD hCS showed strongest overlap with the medial, dorsolateral and orbitofrontal prefrontal cortices (MFC, DFC and OFC) (Fig. 4e). Furthermore, all hCS showed transcriptional similarities with early fetal brain tissue of 8–24 post-conception weeks (Fig. 4f). Treatment and response-status had no significant effect on the spatiotemporal overlap with in vivo brain profiles (Supplementary Fig. 5c–f).
Li-associated genes are involved in sodium ion homeostasis and kidney-related functions
To investigate how Li modulates gene expression, and whether this modulation differs across disease and Li response status groups, six sets of differential expression (DE) analyses were performed as described in the methods. (i) Comparing untreated BD and CTRL hCS identified only 3 DEGs, and of these MLANA was downregulated while USP6 and POTEF were upregulated in BD hCS (FDR < 0.1) (Fig. 5a) (SupplementaryTable 8). (ii) Comparing untreated and Li treated hCS from all 21 donors identified a relatively small number of 132 significant Li-associated DEGs (FDR < 0.1) (Fig. 5b, Supplementary Tables 9, 10 and Supplementary Fig. 6a, b). GO pathway enrichment analysis showed that these genes were enriched for the sodium ion homeostasis pathway and several kidney-related pathways (p < 0.05, FDR < 0.1) (Fig. 5c and Supplementary Table 11). (iii) Comparing untreated and Li treated CTRL hCS identified 74 Li-associated DEGs (Fig. 5d and Supplementary Table 12). The genes were exclusively enriched for the GO term inhibitory synapse assembly (adjust p < 0.05, FDR < 0.1) (Fig. 5f), and the enrichment was mainly driven by upregulation of LGI2, GABRA1 and CBLN4. SynGo analysis showed that these genes were enriched for the synaptic vesicle cellular component (Supplementary Fig. 6c). (iv) Comparing untreated and Li treated BD hCS identified 8 Li-associated DEGs (Fig. 5e). Li upregulated MLANA and CHRM5 and downregulated APOBEC3F, TSPAN9, NFE2, TRABD2B, SMO and ANKRD1 (Fig. 5e). These genes are mainly involved in muscle cell development processes (Fig. 5f), with the main drivers being SMO and ANKRD1. (v) Comparing untreated and treated Li-N hCS identified 29 Li-associated DEGs (FDR < 0.1) (Fig. 5g and Supplementary Table 13). None of the genes were significantly enriched for any GO term. vi) Finally, comparing untreated and treated Li-R hCS identified 5 Li-associated DEGs (FDR < 0.1) (Fig. 5h). Again, MLANA was upregulated, while MYOT, TRDN, TNNI2 and MYL4 were downregulated after Li treatment (Fig. 5h). These genes were significantly enriched for GO terms mostly related to excitable tissue functions like muscle contraction or muscle filament sliding (Fig. 5i). We additionally performed GSEA to further elucidate the affected biological processes, and the results were similar to the GO analysis findings (Supplementary Fig. 6d–f). Notably, we identified three overlapping genes between DEGs identified in this study and BD-associated risk genes [33], i.e. CNTN5, DOCK2 and MAD1L1, which are involved in the formation of axonal connections, actin skeleton remodeling, and cell cycle control, respectively.
a Volcano plot of all DEGs identified for the untreated (Li-) BD vs. CTRL hCS. b Volcano plot of all DEGs identified for the treated (Li+) vs. untreated (Li−) hCS, independent of diagnosis. c Gene ontology (GO) pathway analysis (biological processes) of the 132 Li-associated DEGs identified in all samples independent of diagnosis. d, e Volcano plots of all Li-associated DEGs identified in CTRL and BD hCS, respectively. f GO pathway analysis of the Li-associated DEGs (74) that were unique to CTRL hCS (gray) and DEGs (8) that were unique to BD hCS (blue). g, h Volcano plots of all Li-associated DEGs in Li-N and Li-R hCS, respectively. i GO pathway analysis of the Li-associated DEGs [29] that were unique to Li-N hCS (dark blue) and DEGs [5] that were unique to Li-R hCS (light blue). CTRL (N = 10), Li-N (N = 5), Li-R (N = 6).
Li treatment increases IL-1β secretion in CTRL and BD hCS and decreases TNF-α in CTRL hCS
Systemic levels of the pro-inflammatory cytokines IL-1β, IL-6 and TNF-α are elevated in BD [34,35,36], and some studies suggest Li may have a modulatory effect [37, 38]. Hence, we hypothesized that these cytokines could be increased in supernatants from BD hCS compared to CTRL during differentiation, and that Li treatment would further regulate their levels.
Cytokine profiling of supernatants collected at different time points from day 30 to day 180 showed no difference between BD and CTRL hCS for IL-1β (Fig. 6a, b), but BD hCS had increased TNF-α at day 90 (Fig. 6a) and increased IL-6 at days 160 and 170 compared to CTRL hCS (Fig. 6b). Regression analysis for BD hCS showed a group effect (p = 0.028), and group-by-time interaction (p = 0.041) for TNF-α and group-by-time interaction effect for IL-1β (p = 0.026), highlighting the relevance of these two pro-inflammatory cytokines in BD etiology. Stratifying results based on patient response-status showed decreased IL-6 levels in day 30 Li-N hCS compared to CTRL hCS (Supplementary Fig. 7a), and this trend was reversed after day 60 (Supplementary Fig. 7a). Response status had no consistent effect on basal cytokine levels in the untreated groups at days 160, 170 and 180 (Supplementary Fig. 7b).
a, b IL-1β, IL-6 and TNF-α cytokine levels in hCS at different time points. Li treatment effect in CTRL hCS (c), in BD hCS (d), and stratified in Li-N (e) and Li-R (f). Data is presented as mean ± SEM. g Seahorse oxygen consumption rate (OCR) kinetics graph. h Extracellular acidification rate (ECAR) kinetics graph showing glycolysis activity. i OCR parameters and ECAR basal glycolysis after Li treatment for each condition. j OCR and k ECAR kinetic graphs for Li treatment of BD hCS. l OCR parameters and basal glycolysis after Li treatment in BD hCS. All Seahorse experiments were run in quadruplicates, and values were normalized to total protein. Mitochondrial parameters were normalized to total protein content and data is presented as mean ± SEM. CTRL (N = 10), Li-N (N = 5), Li-R (N = 6). Mann–Whitney U test and Wilcoxon test were used for unpaired and paired group comparisons respectively. *p < 0.05, **p < 0.01.
In CTRL hCS, Li treatment increased IL-1β at day 180, and decreased TNF-α at days 160 and 180 (Fig. 6c). On the other hand, in BD hCS, Li increased IL-1β at days 160, 170 and 180, (Fig. 6d). Li increase of IL-1β in BD hCS seems to be independent of Li-responsiveness, as both Li-N and Li-R showed increased IL-1β at different time points (Fig. 6e, f). No significant difference in IL-6 secretion was detected for any of the treated groups. Interestingly, regression analyses revealed a positive correlation of proinflammatory cytokines with astrocytes and microglia cell fractions, and a negative correlation with neuronal cell types, RGs and OPCs fractions (Supplementary Fig. 7c).
Li increases mitochondrial reserve capacity in BD hCS
Next, we performed mitochondrial characterization of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) (Fig. 6g–l). Untreated Li-N hCS presented decreased maximal respiration capacity compared to CTRL hCS, and Li treatment rescued the normal values in Li-N hCS (Fig. 6i). Moreover, Li treatment for 1 month increased reserve capacity in BD hCS (Fig. 6l), regardless of Li-responsiveness. Interestingly, Li treatment did not alter these parameter in CTRL hCS (Fig. 6i). All analyzed bioenergetics parameters displayed an increasing trend after Li treatment (Fig. 6g–l), and basal glycolysis was significantly increased after treatment when combining all CTRL and BD donors (Supplementary Fig. 7d–f).
Lastly, we previously showed decreased gene intron retention rate (IRR) after 6 h treatment with Li in NPCs, which disappeared after 1 week of treatment [5]. Here, we calculated IRR values for all hCS, and found no significant reduction after 1 month treatment neither in CTRL nor BD hCS (Supplementary Fig. 7g, h), consistent with our finding that Li-induced reduction in IRR occurs during the first hours of treatment and rapidly fades away.
Discussion
Genome-wide association studies (GWAS) have identified 64 risk loci for BD [33]. Risk alleles are enriched in genes involved in synaptic signaling pathways, with high specificity of expression in neurons of the prefrontal cortex and hippocampus [33]. However, there is still limited understanding of which specific biological features are associated with diagnosis, response status and Li treatment. IPSC models used in our experiments may bridge the gap between genotypes and disease phenotypes, as they preserve the genetic signature of patient donors and allow in vitro experimentation with a myriad of conditions and drug treatments.
Treatment impact—effect of duration
A pioneer study showed that 1 week Li treatment regulated 560 genes in human iPSC-derived neurons from Li-R BD patients [3]. Here, after 1 month Li treatment in hCS, we identified a relatively small number of 132 Li-associated DEGs. In our previous studies, Li exposure for 4 days in rats significantly regulated 1108 genes [39], while 6 h treatment in human NPCs regulated 9557 genes, none of which remained significant after 1 week of treatment [5]. Further studies of Li effects in peripheral blood from BD patients found only 56 genes that were regulated after 2–8 weeks treatment [40]. Similarly, reduction in IRR seen after 6 h treatment duration with Li disappeared after 1 week in NPCs [5] and 1 month treatment in our hCS. These results seem to indicate that Li has a substantial short-term impact on gene expression, which in the middle-to-long-term is reversed and homeostasis is restored, likely by compensatory mechanisms or negative-feedback loops, illustrating the heavy influence of treatment duration in Li studies. This duration effect might also explain lack of identification of classical Li-associated genes, such as BCL2, AP1 or GSK3B. Although discrepancy of results with previous studies might be the result not only of biological, but also technical factors [41].
Treatment impact—neuroprotection
Accumulating evidence points to a neuroprotective role of Li in the brain [42,43,44]. Li increases hippocampal and cortical size [31, 32], slows cognitive deterioration [42], preserves cognition in responder patients [43], and its deficiency causes behavioral abnormalities in rats [45]. We found that Li increased CTRL and Li-R hCS size, and increased expression of MAP2 in CTRL hCS and SV2A in both CTRL and Li-N hCS, consistent with findings showing that Li increases MAP2 expression in neural stem cells [46], as well as neurogenesis in animal models [31]. Although most in vitro studies are conducted with cells from patients on Li medication, our results in CTRL hCS suggest that Li neuroprotective effects are independent of diagnosis, in line with findings that long-term low-dose dietary Li intake is associated with longevity in the general population [47, 48].
Electrophysiology and calcium signaling characterization revealed that BD hCS exhibited decreased neuronal excitability and lower neural network activity compared to CTRL hCS. Li treatment increased excitability and rescued decreased currents in BD hCS and had an opposite effect in CTRL hCS. This differs from the findings in previous publications, where increased neuronal excitability and hyperactive neural network activity was found in hippocampal dentate gyrus-like iPSC-derived neurons from BD patients [3, 4]. However, hyperexcitability was only seen in 3-week-old neurons, and Mertens et al. also reported that more mature 8-week-old BD neurons reversed the abnormal excitability to decreased neuron excitability and lower Na+ current amplitudes [3], which is in line with our results. Notably, enhanced accumulation of Li was reported in neurons derived from BD patients compared with CTRL [49], and in dendritic spines Li displaces intracellular Na+ concentrations, thereby reducing its concentration [50]. This accumulation might explain why Li prophylactic action is not immediately reversed upon discontinuation of treatment in BD patients [51]. Moreover, since Li enters neurons primarily through the voltage-gated Na+ channels, it is more likely to affect hyperactive neurons [50].
Treatment impact—side effects
A narrow therapeutic index [52] and several adverse effects restrict Li benefits, and particularly kidney dysfunction is an important contra-indication for long-term Li-prescription [53, 54]. In our DE analysis, we identified several Li-associated DEGs involved in kidney-related processes. Although the enrichment of these pathways was driven by a relatively small number of genes, all of them are expressed at low-medium levels in the brain (https://www.gtexportal.org/). Given that our hCS model contains mainly neurons and glial cells, these findings indicate that regulation of kidney-related genes by Li is systemic and not restricted to the kidneys. While these results concord well with the known side effect burden of Li, their physiological significance in the brain remains unclear.
We also found that Li-associated DEGs are enriched in ion homeostasis pathway. This is consistent with the fact that BD patients under Li treatment are required to monitor sodium chloride (NaCl) intake because any sudden variation of ingested Na+ can affect Li levels. NaCl supplementation diminishes Li side effects [55], and may stabilize serum Li concentration [56], while Li treatment can normalize elevated intracellular Na+ [57].
Etiology insights—ankyrins and Na+ channels dysregulation
Two DEGs associated with diagnosis and Li-treatment in BD hCS, POTEF and ANKRD1 respectively, encode proteins containing ankyrin domains. Variation in genes encoding proteins with ankyrin repeats (TRANK1, SHANK2 and ANK3), is one of the strongest genetic findings in BD [33, 58,59,60]. Ankyrins regulate assembly and anchoring of numerous proteins, including voltage-gated Na+ and K+ ion channels [61,62,63]. Healthy neurons have high densities of voltage-gated ion channels clustered in the axon initial segments (AIS) and nodes of Ranvier [62, 64, 65], suggesting that defects in genes coding for ankyrin proteins might alter neuronal activity in patients. Decreased Na+ currents could be indicative of decreased Na+ channels embedding and clustering, which could affect excitability in BD hCS, including right-shifted IV curves and altered thresholds for currents and APs.
The canonical ankyrins ANK1-3 contain 23-24 ankyrin repeats, but a mapping study of ANK3 fragments demonstrated that repeats 1 to 6 are sufficient for binding Na+ channels [63]. Mutational analysis of these repeats showed a critical role of two lysine residues to bind Nav1.2 [63], which is encoded by another BD-associated gene, SCN2A [33]. Remarkably, POTEF contains 7 ankyrin repeats and has the same critical lysine residues in the same β-hairpin tip between repeats 2-3. Thus, we suggest that upregulated POTEF in untreated BD hCS could reflect a compensatory mechanism to replace defective ankyrins in patients.
Etiology insights—role of pro-inflammatory cytokines
BD pathogenesis is associated with dysregulation of the pro-inflammatory cytokines TNF-α, IL-6 and IL-1β [34,35,36, 66], of which TNF-α and IL-6 are increased during manic and depressive episodes [34, 66]. Here, univariate regression analysis showed a significant increase in TNF-α in BD hCS for all time points. TNF-α regulates apoptotic cascades [67], which may be related to neuronal loss, potentially explaining the reduction in neurons in BD hCS compared to CTRL hCS. Moreover, Li treatment increased IL-1β secretion in both CTRL and BD hCS, in line with most previous studies in immune cells [37, 38, 68]. IL-1β is known to have neurotrophic effects in the mammalian brain, such as neuroprotection [69], enhanced neurogenesis [70] and improved migration of cortical neurons [71], suggesting that the neuroprotective effects of Li may, at least partly, be mediated by IL-1β.
Etiology insights—mitochondrial dysregulation
We previously reported that, in Li-R NPCs, Li treatment increased mitochondrial maximal respiration and reserve capacity [5]. Here, we confirmed that Li significantly increased reserve capacity in BD hCS. However, we also found a deficit of maximal respiration in the Li-N group specifically, and a general effect of Li on mitochondrial function of BD hCS independent of Li responsiveness status. Possible explanations for this discrepancy between our NPCs and hCS findings include the extension of treatment duration from 1 week to 1 month, the greater cell-type complexity of 3D hCS relative to 2D NPCs, and the larger number of donors in the present study.
Limitations
This study has three main limitations. First, the sub-optimal clinical phenotype of patient’s responsiveness to Li, as we were not able to retrospectively confirm lack of Li response in all Li-N donors due to insufficient information in the clinical records. Second, while the hCS model was able to recapitulate key features of human corticogenesis, hCS architecture is rather rudimentary and have a modest electrophysiological activity in comparison to the adult brain cortex. Third, the average age-of-onset of BD is about 25, while gene expression spatiotemporal comparison showed that hCS resemble fetal cortical tissue of 8–24 weeks post-conception, adding a note of caution when interpreting the results. Thus, the disease-related molecular effects captured in the hCS likely stem from predisposing biological factors.
Conclusions
We found that BD hCS had decreased neuronal excitability at the single-cell and neural network levels compared to CTRL hCS, and Li treatment rescued neuronal excitability in BD hCS. Transcriptional profiling revealed that Li treatment regulated genes enriched for biological processes related to sodium ion homeostasis and renal functions. Additionally, Li increased IL-1β protein secretion in CTRL and BD hCS and decreased TNF-α secretion in CTRL hCS. Finally, our findings further support the involvement of mitochondrial regulation in the mechanisms of action of Li. These results illustrate how patient iPSC-derived 3D hCS can be used as a biological model to provide new insight into BD pathophysiology and to advance our understanding of the cellular mechanisms underlying the therapeutic effects of Li.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to national data privacy regulations as they contain information that could compromise research participant privacy and/or consent.
References
Pini S, de Queiroz V, Pagnin D, Pezawas L, Angst J, Cassano GB, et al. Prevalence and burden of bipolar disorders in European countries. Eur Neuropsychopharmacol. 2005;15:425–34.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72.
Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–9.
Stern S, Santos R, Marchetto MC, Mendes APD, Rouleau GA, Biesmans S, et al. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry. 2018;23:1453–65.
Osete JR, Akkouh IA, de Assis DR, Szabo A, Frei E, Hughes T, et al. Lithium increases mitochondrial respiration in iPSC-derived neural precursor cells from lithium responders. Mol Psychiatry. 2021;26:6789–805.
Rybakowski JK, Chlopocka-Wozniak M, Suwalska A. The prophylactic effect of long-term lithium administration in bipolar patients entering treatment in the 1970s and 1980s. Bipolar Disord. 2001;3:63–7.
Wu TN, Lee CS, Wu BJ, Sun HJ, Chang CH, Chen CY, et al. Immunophenotypes associated with bipolar disorder and lithium treatment. Sci Rep. 2019;9:17453.
Yoon SJ, Elahi LS, Pașca AM, Marton RM, Gordon A, Revah O, et al. Reliability of human cortical organoid generation. Nat Methods. 2019;16:75–8.
Kathuria A, Lopez-Lengowski K, Jagtap SS, McPhie D, Perlis RH, Cohen BM, et al. Transcriptomic landscape and functional characterization of induced pluripotent stem cell-derived cerebral organoids in schizophrenia. JAMA Psychiatry. 2020;77:745–54.
Kathuria A, Lopez-Lengowski K, Vater M, McPhie D, Cohen BM, Karmacharya R. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Med. 2020;12:34.
Hibar DP, Westlye LT, Doan NT, Jahanshad N, Cheung JW, Ching CRK, et al. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol Psychiatry. 2018;23:932–42.
Mørch RH, Dieset I, Færden A, Reponen EJ, Hope S, Hoseth EZ, et al. Inflammatory markers are altered in severe mental disorders independent of comorbid cardiometabolic disease risk factors. Psychol Med. 2019;49:1749–57.
Simonsen C, Sundet K, Vaskinn A, Birkenaes AB, Engh JA, Faerden A, et al. Neurocognitive dysfunction in bipolar and schizophrenia spectrum disorders depends on history of psychosis rather than diagnostic group. Schizophr Bull. 2011;37:73–83.
Smith FE, Cousins DA, Thelwall PE, Ferrier IN, Blamire AM. Quantitative lithium magnetic resonance spectroscopy in the normal human brain on a 3 T clinical scanner. Magn Reson Med. 2011;66:945–9.
Squassina A, Meloni A, Congiu D, Bosganas P, Patrinos GP, Lin R, et al. Analysis on in vitro effect of lithium on telomere length in lymphoblastoid cell lines from bipolar disorder patients with different clinical response to long-term lithium treatment. Hum Genomics. 2022;16:45.
Liang KX, Vatne GH, Kristiansen CK, Ievglevskyi O, Kondratskaya E, Glover JC, et al. N-acetylcysteine amide ameliorates mitochondrial dysfunction and reduces oxidative stress in hiPSC-derived dopaminergic neurons with POLG mutation. Exp Neurol. 2021;337:113536.
Szabo A, Akkouh IA, Vandenberghe M, Osete JR, Hughes T, Heine V, et al. A human iPSC-astroglia neurodevelopmental model reveals divergent transcriptomic patterns in schizophrenia. Transl Psychiatry. 2021;11:554.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82.
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9.
Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Chen Y, Lun AT, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284–7.
Abé C, Liberg B, Song J, Bergen SE, Petrovic P, Ekman CJ, et al. Longitudinal cortical thickness changes in bipolar disorder and the relationship to genetic risk, mania, and lithium use. Biol Psychiatry. 2020;87:271–81.
McEvoy GKASoH-SP. AHFS Drug information 2003. Bethesda, Md.: American Society of Health-System Pharmacists; 2003.
Kim JS, Chang MY, Yu IT, Kim JH, Lee SH, Lee YS, et al. Lithium selectively increases neuronal differentiation of hippocampal neural progenitor cells both in vitro and in vivo. J Neurochem. 2004;89:324–36.
Ching CRK, Hibar DP, Gurholt TP, Nunes A, Thomopoulos SI, Abé C, et al. What we learn about bipolar disorder from large-scale neuroimaging: findings and future directions from the ENIGMA Bipolar Disorder Working Group. Hum Brain Mapp. 2022;43:56–82.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Brietzke E, Kapczinski F. TNF-alpha as a molecular target in bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1355–61.
Lu YR, Rao YB, Mou YJ, Chen Y, Lou HF, Zhang Y, et al. High concentrations of serum interleukin-6 and interleukin-8 in patients with bipolar disorder. Medicine. 2019;98:e14419.
Söderlund J, Olsson SK, Samuelsson M, Walther-Jallow L, Johansson C, Erhardt S, et al. Elevation of cerebrospinal fluid interleukin-1ß in bipolar disorder. J Psychiatry Neurosci. 2011;36:114–8.
Liu KJ, Lee YL, Yang YY, Shih NY, Ho CC, Wu YC, et al. Modulation of the development of human monocyte-derived dendritic cells by lithium chloride. J Cell Physiol. 2011;226:424–33.
Petersein C, Sack U, Mergl R, Schönherr J, Schmidt FM, Lichtblau N, et al. Impact of lithium alone and in combination with antidepressants on cytokine production in vitro. J Neural Transm. 2015;122:109–22.
Akkouh IA, Skrede S, Holmgren A, Ersland KM, Hansson L, Bahrami S, et al. Exploring lithium’s transcriptional mechanisms of action in bipolar disorder: a multi-step study. Neuropsychopharmacology. 2020;45:947–55.
Anand A, McClintick JN, Murrell J, Karne H, Nurnberger JI, Edenberg HJ. Effects of Lithium Monotherapy for Bipolar Disorder on Gene Expression in Peripheral Lymphocytes. Mol Neuropsychiatry. 2016;2:115–23.
Forlenza OV, Diniz BS, Radanovic M, Santos FS, Talib LL, Gattaz WF. Disease-modifying properties of long-term lithium treatment for amnestic mild cognitive impairment: randomised controlled trial. Br J Psychiatry. 2011;198:351–6.
Rybakowski JK, Suwalska A. Excellent lithium responders have normal cognitive functions and plasma BDNF levels. Int J Neuropsychopharmacol. 2010;13:617–22.
Malhi GS, Tanious M, Das P, Coulston CM, Berk M. Potential mechanisms of action of lithium in bipolar disorder. Current understanding. CNS Drugs. 2013;27:135–53.
Pickett EE, O’Dell BL. Evidence for dietary essentiality of lithium in the rat. Biol Trace Elem Res. 1992;34:299–319.
Qi L, Tang Y, He W, Pan H, Jiang W, Wang L, et al. Lithium chloride promotes neuronal differentiation of rat neural stem cells and enhances neural regeneration in Parkinson’s disease model. Cytotechnology. 2017;69:277–87.
Zarse K, Terao T, Tian J, Iwata N, Ishii N, Ristow M. Low-dose lithium uptake promotes longevity in humans and metazoans. Eur J Nutr. 2011;50:387–9.
Fajardo VA, LeBlanc PJ, Fajardo VA. Trace lithium in Texas tap water is negatively associated with all-cause mortality and premature death. Appl Physiol Nutr Metab. 2018;43:412–4.
McGhee CE, Yang Z, Guo W, Wu Y, Lyu M, DeLong CJ, et al. DNAzyme-based lithium-selective imaging reveals higher lithium accumulation in bipolar disorder patient-derived neurons. ACS Cent Sci. 2021;7:1809–20.
El-Mallakh RS. Ion homeostasis and the mechanism of action of lithium. Clin Neurosci Res. 2004;4:227–31.
Goodwin FK, Jamison KR. Manic-depressive illness. New zyork, NY, USA: Oxford University Press; 1990;xxi_938-xxi. p.
Geddes JR, Miklowitz DJ. Treatment of bipolar disorder. Lancet. 2013;381:1672–82.
Bocchetta A, Ardau R, Fanni T, Sardu C, Piras D, Pani A, et al. Renal function during long-term lithium treatment: a cross-sectional and longitudinal study. BMC Med. 2015;13:12.
Tondo L, Abramowicz M, Alda M, Bauer M, Bocchetta A, Bolzani L, et al. Long-term lithium treatment in bipolar disorder: effects on glomerular filtration rate and other metabolic parameters. Int J Bipolar Disord. 2017;5:27.
Bleiweiss H. Salts supplements with lithium. Lancet. 1970;1:416.
Tomita T, Goto H, Sumiya K, Yoshida T, Tanaka K, Kohda Y. Stabilization of the serum lithium concentration by regulation of sodium chloride intake: case report. Yakugaku Zasshi. 2016;136:517–21.
Huang X, Lei Z, El-Mallakh RS. Lithium normalizes elevated intracellular sodium. Bipolar Disord. 2007;9:298–300.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Hughes T, Sønderby IE, Polushina T, Hansson L, Holmgren A, Athanasiu L, et al. Elevated expression of a minor isoform of ANK3 is a risk factor for bipolar disorder. Transl Psychiatry. 2018;8:210.
Holmgren A, Hansson L, Bjerkaas-Kjeldal K, Impellizzeri AAR, Gilfillan GD, Djurovic S, et al. Mapping the expression of an ANK3 isoform associated with bipolar disorder in the human brain. Transl Psychiatry. 2022;12:45.
Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–46.
Bréchet A, Fache MP, Brachet A, Ferracci G, Baude A, Irondelle M, et al. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J Cell Biol. 2008;183:1101–14.
Xu M, Cooper EC. An Ankyrin-G N-terminal gate and protein kinase CK2 dually regulate binding of voltage-gated sodium and KCNQ2/3 potassium channels. J Biol Chem. 2015;290:16619–32.
Nelson AD, Jenkins PM. Axonal membranes and their domains: assembly and function of the axon initial segment and node of ranvier. Front Cell Neurosci. 2017;11:136.
Hill AS, Nishino A, Nakajo K, Zhang G, Fineman JR, Selzer ME, et al. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 2008;4:e1000317.
Luo Y, He H, Zhang M, Huang X, Fan N. Altered serum levels of TNF-α, IL-6 and IL-18 in manic, depressive, mixed state of bipolar disorder patients. Psychiatry Res. 2016;244:19–23.
Kim JJ, Lee SB, Park JK, Yoo YD. TNF-alpha-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X(L). Cell Death Differ. 2010;17:1420–34.
Knijff EM, Breunis MN, Kupka RW, de Wit HJ, Ruwhof C, Akkerhuis GW, et al. An imbalance in the production of IL-1beta and IL-6 by monocytes of bipolar patients: restoration by lithium treatment. Bipolar Disord. 2007;9:743–53.
Hewett SJ, Jackman NA, Claycomb RJ. Interleukin-1β in central nervous system injury and repair. Eur J Neurodegener Dis. 2012;1:195–211.
Park SY, Kang MJ, Han JS. Interleukin-1 beta promotes neuronal differentiation through the Wnt5a/RhoA/JNK pathway in cortical neural precursor cells. Mol Brain. 2018;11:39.
Ma L, Li XW, Zhang SJ, Yang F, Zhu GM, Yuan XB, et al. Interleukin-1 beta guides the migration of cortical neurons. J Neuroinflammation. 2014;11:114.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9.
Acknowledgements
The research leading to these results has received funding from the South-Eastern Norway Regional Health Authority (#2022087), the Research Council of Norway (#223273, #248828, #300309), the KG Jebsen Stiftelsen and H2020 Marie Skłodowska-Curie Actions Grant (#80113). Diagram in Fig. 1 was generated with Biorender.com. We also want to acknowledge Martin Falck for his assistance with computational deconvolution, Elin Inderhaug, Kristine Kjeldal and Lars Hansson for their excellent technical assistance, Professor Espen Molden for assistance with drug concentration design, Don Trinh and Mette S. Førsund at Oslo Radium Hospital for slicing organoids, the Norwegian Core Facility for Human Pluripotent Stem Cells Research Centre and the Norwegian Sequencing Centre (www.sequencing.uio.no).
Author information
Authors and Affiliations
Contributions
Conceived and designed the analysis, JRO, IA, OAA and SD; Collected the data, JRO, IA, DRA, OI, MV and TU; Performed the analysis, JRO, IA, DRA, OI, MV, TU and EK; Writing and editing of the paper, JRO, IA, DRA, OI, MV, TU, EK, AS, BH, TH, VMS, OS, SD and OAA; Supervision and funding acquisition, JRO, IA, OAA and SD. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
OAA is a consultant to HealthLytix and has received speaker’s honorarium from Lundbeck and Synovion. SD has received speaker’s honorarium from Lundbeck. The other authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Osete, J.R., Akkouh, I.A., Ievglevskyi, O. et al. Transcriptional and functional effects of lithium in bipolar disorder iPSC-derived cortical spheroids. Mol Psychiatry 28, 3033–3043 (2023). https://doi.org/10.1038/s41380-023-01944-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-01944-0