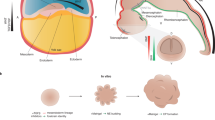
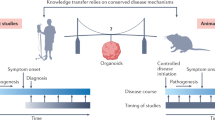
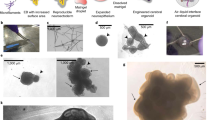
Abstract
Cerebral organoids offer an opportunity to bioengineer experimental avatars of the developing human brain and have already begun garnering relevant insights into complex neurobiological processes and disease. Thus far, investigations into their heterogeneous cellular composition and developmental trajectories have been largely limited to transcriptional readouts. Recent advances in global proteomic technologies have enabled a new range of techniques to explore dynamic and non-overlapping spatiotemporal protein-level programs operational in these humanoid neural structures. Here we discuss these early protein-based studies and their potentially essential role for unraveling critical secreted paracrine signals, processes with poor proteogenomic correlations, or neurodevelopmental proteins requiring post-translational modification for biological activity. Integrating emerging proteomic tools with these faithful human-derived neurodevelopmental models could transform our understanding of complex neural cell phenotypes and neurobiological processes, not exclusively driven by transcriptional regulation. These insights, less accessible by exclusive RNA-based approaches, could reveal new knowledge into human brain development and guide improvements in neural regenerative medicine efforts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327–48.
Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–90.
Fernández V, Llinares‐Benadero C, Borrell V. Cerebral cortex expansion and folding: what have we learned? EMBO J. 2016;35:1021–44.
Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, Riis ES, Freire-Pritchett P, Kelava I, et al. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell. 2021;184:2084–.e19.
Bakken TE, Miller JA, Luo R, Bernard A, Bennett JL, Lee C-K, et al. Spatiotemporal dynamics of the postnatal developing primate brain transcriptome. Hum Mol Genet. 2015;24:4327–39.
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125–1247125.
Tian A, Muffat J, Li Y. Studying human neurodevelopment and diseases using 3D brain organoids. J Neurosci. 2020;40:1186–93.
Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–40.
Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–7.
Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54.
Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–84.
Dong X, Xu SB, Chen X, Tao M, Tang XY, Fang KH, et al. Human cerebral organoids establish subcortical projections in the mouse brain after transplantation. Mol Psychiatry. 2021:26:2964–76.
Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362:eaat6720.
Griffiths JA, Scialdone A, Marioni JC. Using single‐cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol. 2018;14:e8046.
Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol. 2019;58:129–36.
Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17:3369–84.
Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–9.
Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2020;577:537–42.
Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–8.
Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Yang SM, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53.
Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–7.
Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–23.
Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated proteogenomic characterization of human high-grade serous ovarian. Cancer Cell. 2016;166:755–65.
Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62.
Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7.
Djuric U, Brian Lam KH, Kao J, Batruch I, Jevtic S, Papaioannou MD, et al. Defining protein pattern differences among molecular subtypes of diffuse gliomas using mass spectrometry. Mol Cell Proteom. 2019;18:2029–43.
Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature. 2014;509:575–81.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–7.
Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7:10259.
Deeb SJ, Tyanova S, Hummel M, Schmidt-Supprian M, Cox J, Mann M. Machine learning-based classification of diffuse large B-cell lymphoma patients by their protein expression profiles. Mol Cell Proteom. 2015;14:2947–60.
Djuric U, Rodrigues DC, Batruch I, Ellis J, Shannon P, Diamandis P. Spatiotemporal proteomic profiling of human cerebral development. Mol Cell Proteom. 2017;16:1548–62.
Varderidou-Minasian S, Varderidou-Minasian S, Verheijen BM, Schätzle P, Hoogenraad CC, Pasterkamp RJ, et al. Deciphering the proteome dynamics during development of neurons derived from induced pluripotent stem cells. J Proteome Res. 2020;19:2391–403.
Varderidou-Minasian S, Hinz L, Hagemans D, Posthuma D, Altelaar M, Heine VM. Quantitative proteomic analysis of Rett iPSC-derived neuronal progenitors. Mol Autism. 2020;11:1–15.
Singec I, Crain AM, Hou J, Tobe BTD, Talantova M, Winquist AA, et al. Quantitative analysis of human pluripotency and neural specification by in-depth (phospho)proteomic profiling. Stem Cell Rep. 2016;7:527–42.
Benevento M, Tonge PD, Puri MC, Hussein SMI, Cloonan N, Wood DL, et al. Proteome adaptation in cell reprogramming proceeds via distinct transcriptional networks. Nat Commun. 2014;5:1–11.
Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, et al. Self‐organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–29.
Nascimento JM, Saia-Cereda VM, Sartore RC, da Costa RM, Schitine CS, Freitas HR, et al. Human cerebral organoids and fetal brain tissue share proteomic similarities. Front Cell Dev Biol. 2019;7:303.
Goto-Silva L, Ayad NME, Herzog IL, Silva NP, Lamien B, Orlande HRB, et al. Computational fluid dynamic analysis of physical forces playing a role in brain organoid cultures in two different multiplex platforms. BMC Dev Biol. 2019;19:1–10. 2019 191
Dezonne RS, Sartore RC, Nascimento JM, Saia-Cereda VM, Romaõ LF, Alves-Leon SV, et al. Derivation of functional human astrocytes from cerebral organoids. Sci Rep. 2017;7:1–14.
McClure-Begley TD, Ebmeier CC, Ball KE, Jacobsen JR, Kogut I, Bilousova G, et al. Cerebral organoid proteomics reveals signatures of dysregulated cortical development associated with human trisomy 21. BioRxiv. 2018. https://www.biorxiv.org/content/10.1101/315317v2.
Buchsbaum IY, Kielkowski P, Giorgio G, O’Neill AC, Di Giaimo R, Kyrousi C, et al. ECE 2 regulates neurogenesis and neuronal migration during human cortical development. EMBO Rep. 2020;21:e48204.
Chen M, Lee HK, Moo L, Hanlon E, Stein T, Xia W. Common proteomic profiles of induced pluripotent stem cell-derived three-dimensional neurons and brain tissue from Alzheimer patients. J Proteom. 2018;182:21–33.
Lee HK, Velazquez Sanchez C, Chen M, Morin PJ, Wells JM, Hanlon EB, et al. Three dimensional human neuro-spheroid model of Alzheimer’s disease based on differentiated induced pluripotent stem cells. PLoS One. 2016;11:e0163072.
Dakic V, Minardi Nascimento J, Costa Sartore R, MacIel RDM, De Araujo DB, Ribeiro S, et al. Short term changes in the proteome of human cerebral organoids induced by 5-MeO-DMT. Sci Rep. 2017;7:1–13.
Goto-Silva L, Martins M, Murillo JR, Souza LRQ, Vitória G, Oliveira JT, et al. Quantitative profiling of axonal guidance proteins during the differentiation of human neurospheres. Biochim Biophys Acta Proteins Proteom. 2021;1869:140656.
Notaras M, Lodhi A, Barrio-Alonso E, Foord C, Rodrick T, Jones D, et al. Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol Psychiatry. 2021;2021:1–24.
Urresti J, Zhang P, Moran-Losada P, Yu N-K, Negraes PD, Trujillo CA, et al. Cortical organoids model early brain development disrupted by 16p11.2 copy number variants in autism. Mol Psychiatry. 2021;2021:1–21.
Wang X, Liu Q, Zhang B. Leveraging the complementary nature of RNA-Seq and shotgun proteomics data. Proteomics. 2014;14:2676.
Qin X, Sufi J, Vlckova P, Kyriakidou P, Acton SE, Li VSW, et al. Cell-type-specific signaling networks in heterocellular organoids. Nat Methods. 2020;17:335–42.
Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, Lancaster MA. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020. https://doi.org/10.1126/science.aaz5626.
Kelly RT. Single-cell proteomics: progress and prospects. Mol Cell Proteom. 2020;19:1739–48.
Basu S, Campbell HM, Dittel BN, Ray A. Purification of specific cell population by fluorescence activated cell sorting (FACS). J Vis Exp. 2010:10:1546.
Janssens S, Schotsaert M, Manganaro L, Dejosez M, Simon V, García-Sastre A, et al. Facs-mediated isolation of neuronal cell populations from virus-infected human embryonic stem cell–derived cerebral organoid cultures. Curr Protoc Stem Cell Biol. 2019;48:e65.
Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–91.
Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–22. 2014 114.
Lamanna J, Scott EY, Edwards HS, Chamberlain MD, Dryden MDM, Peng J, et al. Digital microfluidic isolation of single cells for -Omics. Nat Commun. 2020;11:1–13.
Berlanda SF, Breitfeld M, Dietsche CL, Dittrich PS. Recent advances in microfluidic technology for bioanalysis and diagnostics. Anal Chem. 2021;93:311–31.
Abe Y, Tada A, Isoyama J, Nagayama S, Yao R, Adachi J, et al. Improved phosphoproteomic analysis for phosphosignaling and active-kinome profiling in Matrigel-embedded spheroids and patient-derived organoids. Sci Rep. 2018;8:11401.
Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–14. 2018 143.
Smith LM, Kelleher NL. Proteoforms as the next proteomics currency: Identifying precise molecular forms of proteins can improve our understanding of function. Science. 2018;359:1106.
Cini FA, Ornelas I, Marcos E, Goto-Silva L, Nascimento J, Ruschi S, et al. d-Lysergic acid diethylamide has major potential as a cognitive enhancer. BioRxiv. 2019. https://doi.org/10.1101/866814.
Author information
Authors and Affiliations
Contributions
All authors contributed equally.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Melliou, S., Sangster, K.T., Djuric, U. et al. The promise of organoids for unraveling the proteomic landscape of the developing human brain. Mol Psychiatry 27, 73–80 (2022). https://doi.org/10.1038/s41380-021-01354-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01354-0