Abstract
Myelodysplastic syndrome with isolated del(5q) is a well-recognized entity with a relatively favorable prognosis. Isolated del(5q) in acute myeloid leukemia is rare and acute myeloid leukemia cases with isolated del(5q) are not well characterized. Del(5q) has been shown to be a poor prognostic marker in acute myeloid leukemia based on multivariable analysis in large cohort studies, which contained mostly cases with del(5q) in the context of multiple chromosomal abnormalities. To further characterize acute myeloid leukemia with isolated del(5q), clinicopathologic characterization including mutation analysis was performed. During a 10-year period, we identified 12 cases of acute myeloid leukemia with isolated del(5q), 7 cases of acute myeloid leukemia with del(5q) plus one additional chromosome abnormality not involving chromosome 7, as well as two control groups composed of 124 cases of acute myeloid leukemia with complex karyotype including del(5q), and 40 cases of myelodysplastic syndrome with isolated del(5q). At diagnosis, cases of acute myeloid leukemia with isolated del(5q) had higher platelet counts (p = 0.044), hemoglobin (p = 0.011), and mean corpuscular volume (p = 0.017) compared with cases of acute myeloid leukemia with complex karyotype including del(5q). Acute myeloid leukemia with isolated del(5q) was less likely therapy-related (p = 0.037), more likely to have IDH1/IDH2 mutations (p = 0.009), and less likely to have TP53 mutations (p = 0.005) when compared to acute myeloid leukemia with complex karyotype including del(5q). Acute myeloid leukemia with isolated del(5q) also showed longer overall survival than acute myeloid leukemia with complex karyotype cases including del(5q) (p = 0.004). In summary, acute myeloid leukemia with isolated del(5q) appeared to show some distinct clinicopathologic and genomic features as compared to cases of acute myeloid leukemia with complex karyotype including del(5q).
Similar content being viewed by others
Introduction
Deletion of chromosome 5q is one of the common genetic abnormalities identified in myeloid neoplasms, especially in myelodysplastic syndrome and acute myeloid leukemia. Based on studies performed mainly on patients with myelodysplastic syndrome, two commonly deleted regions at 5q31 and 5q33 were reported to be minimally necessary for the phenotype; both commonly deleted regions were deleted in most patients with myelodysplastic syndrome with del(5q) [1, 2]. While 5q deletions in myelodysplastic syndrome are often present with additional cytogenetic abnormalities or even in the setting of a complex karyotype [3], myelodysplastic syndrome with isolated del(5q) is a well-recognized entity which demonstrates a relatively favorable prognosis and hallmark features such as macrocytic anemia, variable degree of thrombocytosis, and the presence of hypolobated megakaryocytes [4]. Prior studies have suggested the haploinsufficiency of genes in the commonly deleted regions within the 5q region (such as RPS14 and mir145) play an important role in the pathogenesis of myelodysplastic syndrome with del(5q) [5]. For example, through RNA interference screening, the RPS14 gene has been implicated to block erythroid differentiation [6], while mouse studies demonstrated increased expression of tumor necrosis factor-associated factor-6 (TRAF6), one of the targets of mir145, caused thrombocytosis and the presence of hypolobated megakaryocytes [7].
Myelodysplastic syndrome with isolated del(5q) demonstrates a certain degree of genetic and phenotypic heterogeneity. Recent data have suggested that a single additional cytogenetic abnormality (with the exception of del(7q)/monosomy 7) in cases of myelodysplastic syndrome with del(5q) do not appear to be associated with adverse outcomes [3]. Based on these findings, the entity of myelodysplastic syndrome with isolated del(5q) in the revised 2017 World Health Organization classification now includes cases with one additional cytogenetic abnormality other than del(5q) in the entity, as long as that abnormality does not involve chromosome 7 [8].
In contrast to the relatively favorable outcome observed in myelodysplastic syndrome with isolated del(5q), deletion of 5q has long been considered as a poor prognostic marker in acute myeloid leukemia, as evidenced by multivariable analyses in large-scale acute myeloid leukemia studies investigating the prognostic significance of recurrent chromosome abnormalities [9]. However, as noted in these large cohort studies, only a few acute myeloid leukemia cases demonstrated del(5q) as a sole abnormality, while most of the acute myeloid leukemia cases had del(5q) in the context of additional abnormalities or even as part of a complex karyotype. A previous study focusing on myeloid neoplasms with isolated del(5q) demonstrated the majority of these cases are best classified as myelodysplastic syndrome [10]. In fact, del(5q) was reportedly associated with chromothripsis in acute myeloid leukemia, a signature of genomic instability characterized by extensive genomic rearrangements occurring in a subset of cases with complex karyotype [11]. Isolated del(5q) in acute myeloid leukemia appears to be a rare phenomenon and has not been well characterized.
In this study, we sought to characterize the clinicopathologic features of this rare entity, acute myeloid leukemia with isolated del(5q), with a focus to further elucidate the genetic abnormalities of these neoplasms. We also compared the acute myeloid leukemia cases with isolated del(5q) to acute myeloid leukemia with complex karyotypes including del(5q) to further evaluate the prognostic significance of del(5q) in different genetic backgrounds. Acute myeloid leukemia cases with del(5q) plus single additional chromosome abnormalities (with the exception of chromosome 7 abnormalities) were also collected for comparison.
Materials and methods
Case selection
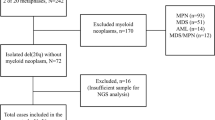
Cases of acute myeloid leukemia with isolated del(5q) and acute myeloid leukemia with del(5q) plus one additional chromosome abnormality were retrieved from the pathology archives at University of Pittsburgh Medical Center from 2008 to 2017. Cases with prior histories of myeloproliferative neoplasms or prior histories of acute myeloid leukemia were excluded from further analyses in all experimental and control groups. Acute myeloid leukemia cases with complex karyotypes including del(5q) and myelodysplastic syndrome with isolated del(5q) diagnosed within the same time period were identified as control groups. Review of the clinical records and pathology material was performed. This study was approved by the University of Pittsburgh Institutional Review Board.
Morphologic evaluation
Morphologic evaluation for dysplasia was performed independently by two hematopathologists (BR and YL) with a previously described method [12]. The overall percentage of dysplasia was quantified in the erythroid, myeloid, and megakaryocytic lineages. Specific features in each lineage were recorded: For erythroid cells, megaloblastoid changes, multinucleation, and nuclear irregularities were evaluated. For myeloid cells, abnormal nuclear shape and hypogranulation were evaluated. For megakaryocytes, micromegakaryocytes, separated nuclear lobes, and hybolobation were evaluated. A minimum of 20 cells in each lineage were required for evaluation; otherwise, the lineage would be scored as “not evaluable”.
Cytogenetic studies
Trypsin-Giemsa banded metaphase cells were obtained from 24 h cultures according to the standard protocol and analyzed at a minimum 400-bands resolution. Results were reported according to the standard International System for Human Cytogenetic Nomenclature [13].
Mutation analysis
DNA samples from the corresponding specimens were subjected to a polymerase chain reaction-based, amplicon target enrichment assay (Illumina TruSeq Amplicon Assay, San Diego, CA). Coding and noncoding regions of the selected genes were enriched and subsequently sequenced on an Illumina MiSeq instrument with paired end, 186 base pair reads. Following mapping of the read data to the human genome (GRCh37/hg19), single nucleotide variants, insertions, and deletions with an allele frequency greater than 5% were identified. FLT3 insertions greater than 15 base pairs were detected to a 0.5% allelic burden. The mutation hotspots of the following genes were interrogated by the test: ABL1, ASXL1, BCOR, BCORL1, BRAF, CALR, CBL, CDKN2A, CSF3R, DNMT3A, ETV6, EZH2, FBXW7, FLT3, GATA2, HRAS, IDH1, IDH2, JAK2, KIT, KRAS, MPL, MYD88, NPM1, NRAS, PHF6, PTEN, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2.
CEBPA mutation analysis was performed by fragment length analysis of the CEBPA gene for screening of the bZIP, TAD1, and TAD2 regions, followed by direct sequencing of positive amplicons utilizing capillary electrophoresis with an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Microarray studies
A microarray study was performed on acute myeloid leukemia with isolated del(5q) case 12 to further delineate the 5q deletion. A custom genome-wide oligonucleotide microarray study using Agilent 4 × 180 K CGH and CGH + SNP designs (Agilent Inc. Santa Clara, CA) was performed on the available DNA sample, as previously described [14].
Statistical analysis
Continuous variables were analyzed using standard nonparametric statistical assays (Kruskal–Wallis, Mann–Whitney) as appropriate. Categorical variables were analyzed using Fisher’s exact test. Survival analysis was performed via the Kaplan–Meier method, and the log-rank test was used to compare survival curves. All statistical analysis was performed using GraphPad Prism Version 8.0.1 (San Diego, CA).
Results
Fifteen cases of acute myeloid leukemia with isolated del(5q) were identified during 2007–2018. Two of these cases (2/15) had prior histories of myeloproliferative neoplasm (polycythemia vera) and one case (1/15) had a prior diagnosis of acute myeloid leukemia with normal karyotype while demonstrating del(5q) during disease progression. These three cases were excluded from further analyses. Eleven of the remaining 12 cases (11/12) presented as de novo acute myeloid leukemia at diagnosis, while one of the 12 cases (1/12) had a prior diagnosis of myelodysplastic syndrome with isolated del(5q). During the same time period, 11 cases of acute myeloid leukemia with del(5q) plus one additional chromosome abnormality which did not involve chromosome 7 were identified. Four of these cases (4/11) with either prior history of myeloproliferative neoplasm (1/11 cases) or acute myeloid leukemia (3/11 cases) were excluded from further analyses. Therefore, study groups comprised 12 cases of acute myeloid leukemia with isolated del(5q) and 7 cases of acute myeloid leukemia with del(5q) plus one additional chromosome abnormality other than chromosome 7.
Two control groups composed of 124 acute myeloid leukemia cases with complex karyotype including del(5q) and 40 cases of myelodysplastic syndrome with isolated del(5q) based on the 2017 World Health Organization classification were identified during the same time period. Laboratory and demographic data from each group were summarized in Table 1 (detailed information for each case listed in Supplementary Table 1). There were no significant differences in the demographics, such as age or male/female ratio, among the four groups. Cases of acute myeloid leukemia with isolated del(5q) had higher hemoglobin level (p = 0.011), platelet count (p = 0.044), and mean corpuscular volume (p = 0.017) at diagnosis when compared with acute myeloid leukemia cases with complex karyotype including del(5q). Furthermore, the acute myeloid leukemia cases with isolated del(5q) were less likely to be therapy-related (p = 0.037) when compared to the acute myeloid leukemia cases with complex karyotype. The hemoglobin level does not appear to be significantly different between the myelodysplastic syndrome and acute myeloid leukemia cases with isolated del(5q) but myelodysplastic syndrome cases with isolated del(5q) demonstrated higher platelet counts at diagnosis (p = 0.002). There were no significant differences in the parameters listed between the acute myeloid leukemia cases with isolated del(5q) and the acute myeloid leukemia cases with del(5q) plus one additional chromosomal abnormality.
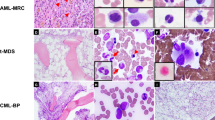
Morphologic review of the acute myeloid leukemia cases with isolated del(5q) revealed some similar and some different morphologic features when compared to myelodysplastic syndrome cases with isolated del(5q). It should be noted that the morphologic evaluation could be limited in the acute myeloid leukemia cases with high blast counts since the remaining hematopoietic elements are markedly decreased. The small hypolobated megakaryocytes invariably identified in the cases of myelodysplastic syndrome with isolated del(5q) were also identified in the majority of acute myeloid leukemia cases with isolated del(5q) (8/8) and the acute myeloid leukemia cases with del(5q) plus one additional chromosomal abnormality (2/3) (Fig. 1, and Supplementary Table 2). The presence of dygranulopoiesis (including hyposegmentation or hypogranularity), which is deemed infrequent in myelodysplastic syndrome with isolated del(5q), was common in acute myeloid leukemia with isolated del(5q) (8/9) and acute myeloid leukemia with del(5q) plus one additional chromosomal abnormality (6/6).
Chromosome analysis of the acute myeloid leukemia cases with isolated del(5q) exhibited a somewhat heterogeneous distribution of breakpoints regarding del(5q) (Fig. 2, and Table 2). All cases of acute myeloid leukemia with isolated del(5q) had deletions involving both commonly deleted regions at 5q31 and 5q33. However, while only 5/35 cases of myelodysplastic syndrome with isolated del(5q) (without any additional chromosome abnormality) demonstrated deletions extending beyond the distal commonly deleted regions within 5q, 7/12 cases of acute myeloid leukemia with isolated del(5q) had deletions extending into the distal commonly retained region beyond 5q33 (p = 0.0054). The most common breakpoint in both acute myeloid leukemia with isolated del(5q) (5/12 cases) and acute myeloid leukemia with del(5q) plus one additional abnormality (4/7 cases) is del(5)(q22q35) while the most common breakpoint for myelodysplastic syndrome cases with isolated del(5q) is del(5)(q13q33), which was identified in 18 of the 40 cases. In the cases with del(5q) plus one additional abnormality, the additional abnormalities consisted of a variety of abnormalities including deletions, trisomies, and translocations (Table 3). While the majority of these cases also demonstrated breakpoints that extended beyond the distal commonly deleted region (4/7 cases) (p = 0.0281 when compared to myelodysplastic syndrome with isolated del(5q)), 1 case showed a 5q deletion that did not span the distal commonly deleted regions at 5q33 and only involved the proximal commonly deleted region at 5q31.
There was heterogeneity in the size/location of the del(5q) in the cases of acute myeloid leukemia with isolated del(5q), acute myeloid leukemia with del(5q) plus one additional abnormality, and myelodysplastic syndrome with isolated del(5q). 5q deletions in the vast majority of myelodysplastic syndrome cases with isolated del(5q) and all acute myeloid leukemia cases with isolated del(5q) involved the commonly deleted regions (gray shading). Acute myeloid leukemia cases with isolated del(5q) and acute myeloid leukemia cases with del(5q) plus one additional abnormality were more likely to show 5q deletions extending into the distal commonly retained region (blue shading) as compared to the cases of myelodysplastic syndrome with isolated del(5q)
Mutation analyses using amplicon target enrichment of coding and noncoding regions of the 37 genes recurrently mutated in myeloid neoplasms were performed on 12 cases of acute myeloid leukemia with isolated del(5q) (Fig. 3). Recurrent mutations in IDH1 (3/12, 25%), IDH2 (4/12, 33%), ASXL1 (4/12, 33%), TP53 (2/12, 17%), and DNMT3A (3/12, 25%) were identified. One third of the acute myeloid leukemia with isolated del(5q) cases (4/12) also demonstrated spliceosomal mutations including mutations in SF3B1 (2/12), SRSF2 (1/12), and U2AF1 (1/12). In comparison, cases of acute myeloid leukemia with complex karyotype including del(5q) demonstrated high percentage of TP53 mutations (11/14, 79%) but low percentage of mutations in IDH1/IDH2 (1/14, 7%) and ASXL1 (0/14). Twenty-one percent of acute myeloid leukemia with complex karyotype including del(5q) showed mutations in spliceosome machinery (3/14). When two groups were compared, cases of acute myeloid leukemia with isolated del(5q) were less likely to carry TP53 mutations (p = 0.005) but more likely to show mutations in IDH1/IDH2 (p = 0.009) and ASXL1 (p = 0.033). The number of mutations in acute myeloid leukemia cases with isolated del(5q) ranged from 0 to 3 (mean: 2.17), while the number of mutations in acute myeloid leukemia cases with complex karyotype including del(5q) ranged from 1 to 4 (mean: 1.57). Intriguingly, one of the acute myeloid leukemia cases with isolated del(5q) demonstrated mutations in NPM1 (allele frequency 36%) and IDH2 (allele frequency 45%); a subsequent microarray study confirmed NPM1 gene located at 5q35 was not deleted in this case (Fig. 4).
Heat map demonstrating the mutational profile of acute myeloid leukemia cases with isolated del(5q) and acute myeloid leukemia cases with complex karyotype including del(5q). Acute myeloid leukemia with isolated del(5q) showed higher percentage of IDH1/IDH2 mutations (p < 0.01), while acute myeloid leukemia with complex karyotype including del(5q) showed higher percentage of TP53 mutations (p < 0.01). Acute myeloid leukemia cases with complex karyotype also demonstrated frequent chromosome 7 abnormalities and del(17p)
Cases of acute myeloid leukemia with isolated del(5q) showed statistically significant longer overall survival when compared with cases of acute myeloid leukemia with complex karyotype including del(5q) (Fig. 5); excluding cases of therapy-related acute myeloid leukemia from the analysis did not alter the statistically significant survival differences. There was no significant difference in survival between cases of acute myeloid leukemia with isolated del(5q) and cases of acute myeloid leukemia with del(5q) plus one additional cytogenetic abnormality. When combining cases of acute myeloid leukemia with isolated del(5q) together with cases of acute myeloid leukemia with del(5q) plus one additional abnormality, the combined group also showed better survival compared with acute myeloid leukemia with complex karyotype including del(5q). Despite the retrospective nature of the study, there was no statistically significant difference in the percentage of cases receiving induction therapy or stem cell transplant among groups of acute myeloid leukemia with isolated del(5q), acute myeloid leukemia with del(5q) plus one additional abnormality, and acute myeloid leukemia with complex karyotype including del(5q). Comparing a group combining the acute myeloid leukemia cases with isolated del(5q) and acute myeloid leukemia cases with del(5q) plus one additional abnormality to acute myeloid leukemia cases with complex karyotype including del(5q) also did not show any statistically significant differences in percentage of the cases receiving induction therapy or stem cell transplant.
a Cases of acute myeloid leukemia with isolated del(5q) had better overall survival compared to cases of acute myeloid leukemia with complex karyotype including del(5q). b Survival differences persisted when acute myeloid leukemia cases with isolated del(5q) and cases with del(5q) plus one additional abnormality were combined together. c The statistically significant better survival in acute myeloid leukemia with isolated del(5q) and d the combined group composed of acute myeloid leukemia with isolated del(5q) and acute myeloid leukemia with del(5q) plus one additional chromosome abnormality persisted after excluding therapy-related lesions from the analysis
Discussion
Del(5q) is a common cytogenetic aberration in myeloid neoplasms; its presence is usually associated with other chromosome abnormalities [3, 9]. Studies regarding myeloid neoplasms with isolated del(5q) are largely on the well-recognized entity of myelodysplastic syndrome with isolated del(5q). The 5q deletions in myeloid neoplasms usually involve both commonly deleted regions at 5q31 and 5q33 [1, 2], as seen in all of our acute myeloid leukemia cases with isolated del(5q). In our cohort, the 5q deletions in acute myeloid leukemia with isolated del(5q) were more likely to extend beyond the distal commonly deleted regions when compared with myelodysplastic syndrome with isolated del(5q). The phenomenon that 5q deletions in acute myeloid leukemia tend to involve the regions commonly retained in myelodysplastic syndrome was also described in a prior study including many cases with additional chromosome abnormalities besides del(5q) [2]. One interesting but unusual acute myeloid leukemia case with isolated del(5q) in our cohort showed a concurrent NPM1 mutation, presumptively in high percentage of neoplastic cells with an allele frequency of 36%. A previous study suggested the NPM1 gene, located within the commonly retained region distal to the commonly deleted region within 5q, was usually deleted in high-grade myelodysplastic syndrome or acute myeloid leukemia [15]. It was also suggested that acute myeloid leukemia cases with uncommonly smaller 5q deletions sparing the commonly retained regions may demonstrate mutations in MAML1 and/or NPM1 (both located within the 5q commonly retained regions) [2]. Although the chromosome analysis used in our study does not necessarily provide the best resolution and may potentially miss significant microdeletions and loss of heterozygosity, our findings based on the karyotyping and the presence of a case with concurrent NPM1 mutation suggest the importance of genes outside the known commonly deleted regions in leukemogenesis.
The mutational landscape of myelodysplastic syndrome with isolated del(5q) was characterized in several prior studies that described mutations in TP53, ASXL1, TET2, DNMT3A, and SF3B1 [16, 17]. TP53 mutations, reported in roughly 18% of the patients of myelodysplastic syndrome with isolated del(5q), were reported to predict disease progression and were associated with lack of complete cytogenetic response to lenalidomide [18,19,20]. Given the association between TP53 mutation and leukemia progression in myelodysplastic syndrome with isolated del(5q), it is noteworthy that the incidence of TP53 mutation in acute myeloid leukemia with isolated del(5q) is not significantly higher than that reported in myelodysplastic syndrome with isolated del(5q). However, it is not unanticipated that the incidence of TP53 mutations in acute myeloid leukemia with isolated del(5q) was lower than that in acute myeloid leukemia with complex karyotype including del(5q) since a high percentage of TP53 alterations was reported in acute myeloid leukemia cases with complex karyotype in prior studies [11, 21, 22]. Although no IDH1/IDH2 mutations have been reported in myelodysplastic syndrome with isolated del(5q) based on the published data from a total of 100 patients tested [16, 23], we uncovered a high percentage of IDH1/IDH2 mutations in acute myeloid leukemia with isolated del(5q). Suggested by several prior studies, mutations of IDH1/IDH2 could block differentiation in hematopoietic cells, which can be reversed by pharmacologic inhibition [24,25,26]. IDH1/IDH2 mutations have previously been linked to leukemic transformation in MPN [27, 28] and myelodysplastic syndrome [29, 30]. A prior case series has described the presence of IDH1/IDH2 mutations in four high-grade myelodysplastic syndrome and two acute myeloid leukemia with isolated del(5q) [31]. Our findings of frequent IDH1/IDH2 mutations in acute myeloid leukemia with isolated del(5q) suggests that in the context of haploinsufficiency of genes within the deleted 5q region, the differentiation block induced by IDH1/IDH2 mutation may contribute to an acute leukemic phenotype.
Although del(5q) has long been considered a poor prognostic factor in acute myeloid leukemia, we hypothesized the common co-occurrence of del(5q) with multiple other cytogenetic abnormalities may confound the assessment of its significance and its prognostic value may depend on the overall genetic backgrounds. Our study demonstrated better overall survival in acute myeloid leukemia with isolated del(5q) compared to acute myeloid leukemia cases with complex karyotype including del(5q), even after excluding therapy-related lesions. The only prior study that alluded to survival in acute myeloid leukemia with isolated del(5q) used cases of acute myeloid leukemia with isolated del(5q) as part of the control group and demonstrated myelodysplastic syndrome with isolated del(5q) had better survival compared with all other myeloid neoplasms with isolated del(5q) combined together [10]. Our findings are particularly compelling since we underscored the different prognostic value of del(5q) even in the same high-grade disease entity such as acute myeloid leukemia by choosing acute myeloid leukemia with complex karyotype including del(5q) as the control group. Considering World Health Organization classification, though it is difficult to directly compare data among different studies, acute myeloid leukemia cases with isolated del(5q) in our study appeared to demonstrate a comparable overall survival with acute myeloid leukemia, NOS and a trend toward better survival than acute myeloid leukemia with myelodysplasia-related changes when compared with previously published studies [32, 33]. Larger-scale studies will be of interest to confirm the findings of our study.
Acute myeloid leukemia with isolated del(5q) appeared to show some clinicopathologic findings similar to those seen in myelodysplastic syndrome with isolated del(5q) based on our data, such as higher platelet counts and the presence of small, hypolobated megakaryocytes. This result is not surprising since both cases of acute myeloid leukemia and myelodysplastic syndrome with isolated del(5q) shared common genetic abnormalities involving 5q, including the haploinsufficiency of RPS14 and deletion of mir145/mir146. These abnormalities have been shown to contribute to the hallmark features in myelodysplastic syndrome with isolated del(5q) through their impact on erythropoiesis and megakaryopoiesis [6, 7, 34]. Nonetheless, like myelodysplastic syndrome cases with isolated del(5q), acute myeloid leukemia lesions with isolated del(5q) also demonstrate phenotypic and genetic heterogeneity.
Regarding the limitations of the study, we acknowledge it is difficult to completely exclude the possibility that patients with acute myeloid leukemia with isolated del(5q) may have had clinicopathologic findings that qualified for a diagnosis of myelodysplastic syndrome prior to their acute myeloid leukemia diagnoses, although all but one of our cases presented in a de novo fashion. In addition, we did not identify clear differences in survival between acute myeloid leukemia cases with isolated del(5q) and acute myeloid leukemia cases with del(5q) plus one additional chromosome abnormality, but the low number of cases in both rare entities may limit the statistical analyses.
In summary, we showed acute myeloid leukemia cases with isolated del(5q) had better survival than acute myeloid leukemia cases with del(5q) as part of a complex karyotype. The prognostic value of del(5q) did not appear to be identical in different genetic backgrounds even in the entity of acute myeloid leukemia. Acute myeloid leukemia with isolated del(5q) also appeared to show a high percentage of IDH1/IDH2 mutations, while it demonstrated a similar incidence of TP53 mutations compared to myelodysplastic syndrome with isolated del(5q) and a much lower incidence of TP53 mutations compared to acute myeloid leukemia with complex karyotype including del(5q). Our data suggest that acute myeloid leukemia with isolated del(5q) has some distinct clinicopathologic and genetic features compared to cases of acute myeloid leukemia with complex karyotype including del(5q).
References
Sole F, Espinet B, Sanz GF, Cervera J, Calasanz MJ, Luno E, et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo Cooperativo Espanol de Citogenetica Hematologica. Br J Haematol. 2000;108:346–56.
Jerez A, Gondek LP, Jankowska AM, Makishima H, Przychodzen B, Tiu RV, et al. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J Clin Oncol. 2012;30:1343–9.
Mallo M, Cervera J, Schanz J, Such E, Garcia-Manero G, Luno E, et al. Impact of adjunct cytogenetic abnormalities for prognostic stratification in patients with myelodysplastic syndrome and deletion 5q. Leukemia. 2011;25:110–20.
Hasserjian RP, Le Beau MM, List AF, Bennett JM, Brunning RD, Thiele J. Myelodysplastic syndrome with isolated del(5q). In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri S, Stein H, et al. editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised. 4th ed. Lyon: International Agency for Research on Cancer; 2017. p. 115–6.
Ebert BL. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Semin Oncol. 2011;38:621–6.
Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–9.
Ribezzo F, Snoeren IAM, Ziegler S, Stoelben J, Olofsen PA, Henic A, et al. Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia. 2019;33:1759–72.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65.
Patnaik MM, Lasho TL, Finke CM, Knudson RA, Ketterling RP, Chen D, et al. Isolated del(5q) in myeloid malignancies: clinicopathologic and molecular features in 143 consecutive patients. Am J Hematol. 2011;86:393–8.
Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–21.
Weinberg OK, Gibson CJ, Blonquist TM, Neuberg D, Pozdnyakova O, Kuo F, et al. Association of mutations with morphological dysplasia in de novo acute myeloid leukemia without 2016 WHO Classification-defined cytogenetic abnormalities. Haematologica. 2018;103:626–33.
McGowan-Jordan J, Simons A and Schmid M. ISCN 2016: an international system for human cytogenomic nomenclature. Basel: Karger; 2016.
Peterson JF, Aggarwal N, Smith CA, Gollin SM, Surti U, Rajkovic A, et al. Integration of microarray analysis into the clinical diagnosis of hematological malignancies: How much can we improve cytogenetic testing? Oncotarget. 2015;6:18845–62.
La Starza R, Matteucci C, Gorello P, Brandimarte L, Pierini V, Crescenzi B, et al. NPM1 deletion is associated with gross chromosomal rearrangements in leukemia. PLoS ONE. 2010;5:e12855.
Fernandez-Mercado M, Burns A, Pellagatti A, Giagounidis A, Germing U, Agirre X, et al. Targeted re-sequencing analysis of 25 genes commonly mutated in myeloid disorders in del(5q) myelodysplastic syndromes. Haematologica. 2013;98:1856–64.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. quiz 99
Jadersten M, Saft L, Smith A, Kulasekararaj A, Pomplun S, Gohring G, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–9.
Mallo M, Del Rey M, Ibanez M, Calasanz MJ, Arenillas L, Larrayoz MJ, et al. Response to lenalidomide in myelodysplastic syndromes with del(5q): influence of cytogenetics and mutations. Br J Haematol. 2013;162:74–86.
Mossner M, Jann JC, Nowak D, Platzbecker U, Giagounidis A, Gotze K, et al. Prevalence, clonal dynamics and clinical impact of TP53 mutations in patients with myelodysplastic syndrome with isolated deletion (5q) treated with lenalidomide: results from a prospective multicenter study of the german myelodysplastic syndrome study group (Gmyelodysplastic syndrome). Leukemia. 2016;30:1956–9.
Sebaa A, Ades L, Baran-Marzack F, Mozziconacci MJ, Penther D, Dobbelstein S, et al. Incidence of 17p deletions and TP53 mutation in myelodysplastic syndrome and acute myeloid leukemia with 5q deletion. Genes Chromosomes Cancer. 2012;51:1086–92.
Mrozek K, Eisfeld AK, Kohlschmidt J, Carroll AJ, Walker CJ, Nicolet D, et al. Complex karyotype in de novo acute myeloid leukemia: typical and atypical subtypes differ molecularly and clinically. Leukemia. 2019;33:1620–34.
Patnaik MM, Lasho TL, Finke CM, Gangat N, Caramazza D, Holtan SG, et al. WHO-defined ‘myelodysplastic syndrome with isolated del(5q)’ in 88 consecutive patients: survival data, leukemic transformation rates and prevalence of JAK2, MPL and IDH mutations. Leukemia. 2010;24:1283–9.
DiNardo CD. Ivosidenib in IDH1-mutated acute myeloid leukemia. N Engl J Med. 2018;379:1186.
Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–41.
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–31.
Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–70.
Pardanani A, Lasho TL, Finke CM, Mai M, McClure RF, Tefferi A. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia. 2010;24:1146–51.
DiNardo CD, Jabbour E, Ravandi F, Takahashi K, Daver N, Routbort M, et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia. 2016;30:980–4.
Thol F, Weissinger EM, Krauter J, Wagner K, Damm F, Wichmann M, et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica. 2010;95:1668–74.
Pardanani A, Patnaik MM, Lasho TL, Mai M, Knudson RA, Finke C, et al. Recurrent IDH mutations in high-risk myelodysplastic syndrome or acute myeloid leukemia with isolated del(5q). Leukemia. 2010;24:1370–2.
Weinberg OK, Seetharam M, Ren L, Seo K, Ma L, Merker JD, et al. Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood. 2009;113:1906–8.
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31.
Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Rea, B., Aggarwal, N., Yatsenko, S.A. et al. Acute myeloid leukemia with isolated del(5q) is associated with IDH1/IDH2 mutations and better prognosis when compared to acute myeloid leukemia with complex karyotype including del(5q). Mod Pathol 33, 566–575 (2020). https://doi.org/10.1038/s41379-019-0396-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0396-4