Abstract
DNA mismatch repair protein deficient colon cancer frequently displays reduced CDX2 expression, and recent literature has suggested that negative CDX2 expression is a poor prognostic biomarker in colon cancer. We have recently demonstrated that SATB2 is an immunohistochemical marker that is complementary to CDX2. Using a tissue microarray approach, we evaluated SATB2 and CDX2 immunohistochemical expression in 514 patients with colonic adenocarcinoma including 146 with mismatch repair protein deficient tumors and correlated expression with histopathologic variables, molecular alterations, and survival. Overall, SATB2-negative and/or CDX2-negative expression was identified in 33% of mismatch repair protein deficient tumors compared with only 15% of mismatch repair protein proficient tumors (p < 0.001) and in 36% of BRAF V600E mutated compared with only 13% of BRAF wild-type tumors (p < 0.001). Both SATB2-negative and CDX2-negative colonic adenocarcinomas more often displayed lymphatic invasion, venous invasion, and perineural invasion (all with p < 0.05). SATB2-negative expression was also more frequently identified in tumors with mucinous or signet ring cell differentiation (p < 0.01 for both). In a multivariable analysis of survival in patients with mismatch repair protein deficient tumors (n = 131), only tumor stage (p = 0.01) and SATB2-negative and/or CDX2-negative expression (p = 0.009) independently predicted disease-specific survival. Of the 99 patients with stage II or III mismatch repair protein deficient tumors, death from disease only occurred in patients with either SATB2-negative or CDX2-negative tumors, and no patients with SATB2-positive/CDX2-positive tumors developed recurrence or died of disease. SATB2 and CDX2 expression had no effect on patient survival in mismatch repair protein proficient, BRAF-mutated, or KRAS-mutated tumors. In summary, our results suggest that SATB2 and CDX2 are prognostic biomarkers in patients with mismatch repair protein deficient colon cancer and that inclusion of SATB2 and CDX2 immunohistochemistry may be helpful as part of a comprehensive pathologic risk assessment in mismatch repair protein deficient colon cancer.
Similar content being viewed by others
Introduction
An estimated 140,000 individuals are diagnosed with colorectal carcinoma each year, and ~50,000 will die from this disease, making colorectal carcinoma the third leading cause of cancer deaths in the United States [1]. The treatment and prognosis of colorectal carcinoma is determined primarily by TNM staging but significant prognostic heterogeneity remains within each stage category, particularly in stage II and stage III carcinomas [2]. For example, in stage II colon cancer the 5-year survival after complete surgical resection ranges between 65 and 85% [3]. The wide range of survival calls for robust prognostic markers that can identify patients with an increased risk of disease recurrence and death from disease. To date, markers identified for poor prognosis include certain histopathologic variables and molecular alterations such as BRAF V600E mutation in the setting of proficient DNA mismatch repair protein expression/microsatellite stability [4,5,6] while mismatch repair protein deficiency/high-level microsatellite instability has generally been associated with improved survival [4, 5, 7,8,9,10,11,12]. However, a subset of patients with mismatch repair protein deficient tumors can develop relapse and die of disease. This suggests additional biomarkers are needed to stratify patients with mismatch repair protein deficient tumors into prognostic groups.
Both the caudal type homeobox transcription factor 2 (CDX2) and the special AT-rich sequence binding protein (SATB2) are immunohistochemistry markers for normal colorectal epithelium and colorectal carcinoma. CDX2 has been identified as a prognostic biomarker in colonic adenocarcinoma; patients with CDX2-negative adenocarcinomas have reduced survival [13,14,15]. SATB2 is a transcription factor regulating chromatin remodeling and transcription [16]. It has only recently been identified as a tissue type-specific protein in the gastrointestinal tract with selective expression in benign and neoplastic epithelial cells of colorectal origin [17]. SATB2 has been shown to be complementary to CDX2 as a diagnostic immunohistochemistry biomarker [17,18,19,20,21,22,23]. Limited literature data suggest that SATB2 suppresses the progression of colorectal carcinoma in vitro and in vivo [24] and that patients with SATB2-negative colonic adenocarcinomas have reduced survival [25, 26].
Loss of CDX2 expression in mismatch repair protein deficient colorectal carcinoma has been well-documented in the literature [27,28,29,30]. Loss of SATB2 expression in mismatch repair protein deficient colorectal carcinoma has been recently reported by Eberhard et al. [26] and in an analysis by our group [18]. In particular, in a survey of 246 colorectal carcinomas, we demonstrated that, similar to CDX2, loss of SATB2 expression is also preferentially seen in colorectal carcinomas with mismatch repair protein deficiency [18]. In addition, while most mismatch repair protein proficient colorectal carcinomas exhibit dual positive expression of SATB2 and CDX2 (SATB2+/CDX2+), half of the mismatch repair protein deficient colorectal carcinomas demonstrate negative expression of either SATB2 or CDX2 (SATB2−/CDX2+, SATB2+/CDX2−) or both (SATB2−/CDX2−) [18].
Based on these findings, we hypothesized that loss of either SATB2 or CDX2 immunohistochemical expression is associated with poor prognosis in mismatch repair deficient colonic adenocarcinoma. To test this hypothesis, we assessed the association between SATB2 and CDX2 immunohistochemistry expression with mismatch repair protein status, status of KRAS and BRAF mutations, histopathologic variables associated with poor prognosis, and patient survival in 514 colon cancers collected from two tertiary care medical centers.
Materials and methods
Study group
The clinicopathologic records of 514 patients with surgically resected primary colonic adenocarcinoma were reviewed, including 302 resected at the University of Pittsburgh Medical Center and 212 resected at Stanford University Medical Center. The 302 colonic adenocarcinomas from University of Pittsburgh Medical Center included a consecutive series of 191 mismatch repair protein proficient colonic adenocarcinomas resected between 2011 and 2012 and a consecutive series of 111 mismatch repair protein deficient colonic adenocarcinomas resected between 2010 and 2015. The 212 colonic adenocarcinomas from Stanford University Medical Center were consecutively resected between 2005 and 2009, and SATB2 and CDX2 expression and mismatch repair protein status in tumors from these patients have been previously reported by our group [18]. Patients with rectal adenocarcinomas were specifically excluded given their different therapeutic management compared with colonic adenocarcinoma. The type of initial surgical procedure, extent of disease, demographic information, and clinical follow-up were obtained from medical records under the guidelines of the Institutional Review Board (PR016040136).
For each tumor, representative areas of the tumor were selected for tissue microarrays from paraffin blocks based on hematoxylin and eosin-stained sections. Each tumor was represented by two or three cores on the tissue microarray. For tumors with more than one morphologic growth pattern, tissue cores were selected to include all morphologic patterns present within an individual tumor. The tissue microarrays were created using 1.0 mm tissue cores using a tissue arrayer (Beecher Instruments, Silver Spring, MD) according to a previously described method [31].
Immunohistochemical analysis
SATB2 (clone EP281, Cell Marque, Rocklin, CA) and CDX2 (clone CDX2-88, Biogenex, Fremont, CA) immunohistochemical stains were performed on all available tumor samples. Mismatch repair protein immunohistochemistry was performed using primary monoclonal antibodies against MLH1 (Stanford: clone G168-728, Ventana, Tucson, AZ; University of Pittsburgh Medical Center: clone M1, Ventana), MSH2 (Stanford: clone FE11, Oncogene Research Products, Cambridge, MA; University of Pittsburgh Medical Center: clone G219-1129, Ventana), MSH6 (Stanford: clone 44, BD Biosciences, San Jose, CA; University of Pittsburgh Medical Center: clone 44, Ventana), and PMS2 (Stanford: clone A16-4, BD Biosciences; University of Pittsburgh Medical Center: clone EPR3947, Cell Marque, Rocklin, CA).
SATB2 and CDX2 were scored semiquantitatively for both staining intensity and proportion of tumor cells with nuclear staining. Staining intensity was scored as 0, absent; 1+, weak nuclear staining visible with ×10 or greater objective magnification; 2+, strong nuclear staining visible at ×2 or ×4 objective magnification. Percentage of tumor cells with nuclear staining was estimated by visual inspection in increments of 5%. A modified H-score for SATB2 and CDX2, defined as intensity score × percentage of tumor cells with nuclear staining, was calculated. Positive SATB2 and CDX2 results were defined as nuclear staining in tumor cells, regardless of either staining intensity or the proportion of tumor cells with nuclear staining.
For MLH1, PMS2, MSH2, and MSH6, preserved expression was defined as nuclear staining within tumor cells, using infiltrating lymphocytes as a positive internal control. Loss of protein expression for MLH1, PMS2, MSH2, and MSH6 was defined as complete absence of nuclear staining within tumor cells with concurrent positive labeling in internal non-neoplastic tissues. Proficient mismatch repair protein expression was defined as preserved nuclear expression of all four mismatch repair proteins. Deficient mismatch repair protein expression was defined as loss of protein expression of at least one of the four mismatch repair proteins. For tumors resected at the University of Pittsburgh Medical Center, mismatch repair protein immunohistochemistry was performed on whole tumor sections at the time of initial pathologic assessment as part of routine evaluation, and all mismatch repair protein results were confirmed by re-review of the mismatch repair protein immunohistochemical stains. For tumors resected at Stanford University, mismatch repair protein immunohistochemistry was performed on the tissue microarrays with any tumor demonstrating loss of mismatch repair protein expression on the tissue microarray examined by whole tissue sections of the tumor to confirm loss of mismatch repair protein expression.
To validate the SATB2 and CDX2 immunohistochemistry results in mismatch repair protein deficient tumors from the tissue microarrays, mismatch repair protein deficient colon cancers with negative SATB2 expression (n = 11) or negative CDX2 expression (n = 26) with available tumor blocks were re-evaluated by immunohistochemistry on whole tumor sections. Whole tumor section immunohistochemistry confirmed the tissue microarray results in 34 of 37 (92%) cases. Negative SATB2 expression was confirmed in all 11 tested cases with internal positive control expression in non-neoplastic colonic crypt epithelium. For the 26 tumors with negative CDX2 expression on the tissue microarray analysis, evaluation of whole tissue sections confirmed the negative expression of CDX2 in 23 (88%) tumors with internal control positive expression in non-neoplastic colonic crypts. In remaining three tumors, heterogenous and weak CDX2 expression in a minority of tumor cells was identified on whole section immunohistochemistry, and these three tumors were reclassified as having CDX2-positive expression given the whole tissue section analysis.
Pathologic evaluation
Histologic examination of all pathology slides from 362 (70%) cases was performed and the following histologic features were re-evaluated for each case: histologic grade, stage, mucinous component, signet ring cell component, lymphatic invasion, venous invasion, perineural invasion, tumor infiltrating lymphocytes, and tumor budding. All histologic features were assessed using routine hematoxylin and eosin-stained slides. Tumor location was stratified into three categories: cecum, right colon (which encompasses the ascending colon, hepatic flexure, and transverse colon), and left colon (which encompasses the splenic flexure, descending colon, and sigmoid colon).
A two-tier tumor grading scheme was employed. Low-grade was defined as > 50% gland formation. High-grade was defined as <50% gland formation and/or the presence of signet ring cell differentiation. The presence of tumor infiltrating lymphocytes was assessed using the criteria outlined by Greenson et al. [32]. Briefly, each histologic section of tumor was assessed at low-power (×2 or ×4) objective magnification to identify the “hotspot” area with the most tumor infiltrating lymphocytes. In the “hotspot” area, the number of tumor infiltrating lymphocytes was counted in five consecutive high-power (×40 objective) fields. If on average ≥3 lymphocytes per high-power field were identified within the tumor, the tumor was scored as positive for tumor infiltrating lymphocytes. If on average < 3 lymphocytes per high-power field within the tumor were identified, the tumor was scored as negative for tumor infiltrating lymphocytes. Tumor budding was assessed using the method advocated by the International Tumor Budding Consensus Conference [33] and adopted by the College of American Pathologists colorectal carcinoma protocol [34]. Tumor buds were defined as isolated cancer cells or a cluster of < 5 neoplastic cells at the invasive front of the tumor. The tumor invasive front was assessed at a scanning (×10 objective) magnification for the area with maximal tumor budding. In this area, the number of tumor buds was determined at the invasive front in a 0.785 mm2 area. Tumors were classified using the College of American Pathologists colorectal carcinoma protocol three-tier scheme as follows: low tumor budding if 0 to 4 tumor buds were identified per 0.785 mm2, intermediate tumor budding if 5–9 tumor buds were identified per 0.785 mm2, and high tumor budding if ≥ 10 tumor buds were identified per 0.785 mm2 [34].
KRAS and BRAF V600E mutational analysis
For tumors from Stanford University Medical Center, tumor targets were manually microdissected from 4-micron unstained histologic sections. DNA was extracted from paraffin sections, using the DNeasy tissue kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. Detection of BRAF mutations was performed as previously described [35]. Briefly, real-time polymerase chain reaction was performed using allele specific primers designed to selectively amplify the wild-type (T1799) and mutant (A1799) BRAF alleles, i.e., to detect BRAF c.1799T>A (V600E) mutation. After amplification, samples were subjected to a temperature ramp from 60 to 99 °C, increasing 1 °C at each step. For wild-type samples, single peaks were observed at 80 °C, whereas samples containing mutant alleles produced single peaks at 85 °C. Detection of KRAS mutations were performed as previously described [35], using a validated KRAS mutation kit (Entrogen, Tarzana, CA) that identifies the common somatic mutations located in codons 12, 13, and 61 of the KRAS gene. Molecular testing for tumors resected at Stanford University was performed after the initial pathologic evaluation and was not part of routine evaluation.
For tumors from the University of Pittsburgh Medical Center, detection of BRAF mutations was performed using real-time polymerase chain reaction and post-polymerase chain reaction fluorescence melting curve analysis on LightCycler (Roche Applied Science, Indianapolis, IN), as previously described [36]. This assay detects BRAF mutations in codons 599, 600, and 601. However, only BRAF mutation status at codon 600 which corresponds to the c.1799T>A (V600E) mutation was correlated with status of CDX2 and SATB2 expression and patient survival. Detection of KRAS gene mutations in exon 2 (codons 12 and 13) and exon 3 (codon 61) were performed as previously described [6]. Molecular testing for tumors resected at the University of Pittsburgh Medical Center was performed at the time of initial pathologic assessment as part of routine evaluation.
Colon cancer data in the Cancer Genome Atlas Pan Cancer Atlas
Normalized Illumina HiSeq mRNA expression data of SATB2 and CDX2, microsatellite status, and status of KRAS and BRAF mutations in the colon cancer dataset in the Cancer Genome Pan Cancer Atlas were obtained by searching and downloading through the cBioPortal for Cancer Genomics [37, 38]. Clinical follow-up data were obtained from the NCI Genomic Data Commons (GDC) [39]. mRNA expression levels of SATB2 and CDX2 were stratified by microsatellite status, the presence of BRAF V600E, and the presence of KRAS exon 2 and/or exon 3 mutations.
Statistical analysis
Chi-square or Fisher exact tests were used to characterize the relationship between categorical variables, as appropriate. Kruskal–Wallis tests were used to characterize the relationship between continuous variables. Disease-specific survival and disease-free survival were the primary endpoints. Death occurring within one month of the initial operation was attributed to operative mortality and was not included in the survival analysis. Disease-specific survival was defined as the time (measured in months) from the date of initial diagnosis to the date of death from disease and censored at the date of last clinical follow-up or death from an unrelated cause. Disease-free survival was defined as the time (measured in months) from the date of initial diagnosis to the date of distant disease recurrence (i.e., tumor involving the peritoneum and/or other organs) and censored at the date of last clinical follow-up or death. Patients with stage IV disease at initial presentation were not included in the disease-free survival analysis. Survival rates were determined by the Kaplan–Meier method and differences between groups were evaluated by log-rank test. Hazard ratios were calculated from a Cox proportional hazard model to identify individual predictors of disease-specific survival. Multivariate analysis of significant individual risk factors (p < 0.05) was performed using Cox proportional hazard regression to identify independent risk factors for disease-specific survival. Data from univariate and multivariate analyses were reported as hazard ratios with 95% confidence intervals. All statistics were assessed using two-sided tests with p-values < 0.05 considered statistically significant. Statistical analyses were performed using SPSS (for Windows 23, IBM, Armonk, NY) and GraphPad Prism 7 for Windows Version 7.00 (GraphPad Software, La Jolla, CA).
Results
Negative SATB2 and CDX2 expression is associated with adverse histopathologic features in colonic adenocarcinoma
Of the 514 colonic adenocarcinomas included in the tissue microarrays (Table 1), 499 had adequate tumor tissue available to assess SATB2 expression. Sixty-seven of 499 tumors (13%) had negative SATB2 immunohistochemical expression (Fig. 1). Histologic review of all pathology slides was performed in 357 (72%) of 499 tumors. Compared with SATB2-positive tumors, SATB2-negative tumors more often displayed adverse histologic features including high tumor grade (41% vs. 11%, p < 0.001), lymphatic invasion (68% vs. 51%, p = 0.04), venous invasion (39% vs 22%, p = 0.02), and perineural invasion (36% vs. 22%, p = 0.04). Patients with SATB2-negative tumors more frequently had tumors located in the cecum and right colon (78%) compared with patients with positive SATB2-positive tumors (59%) (p = 0.01). Mucinous and signet ring cell differentiation were also more commonly observed in SATB2-negative tumors compared with SATB2-positive tumors (mucinous: 50% vs. 25%, p = 0.002; signet ring cell: 16% vs. 3%, p < 0.001).
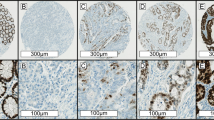
Examples of colonic adenocarcinomas with loss of SATB2 (a, b) and CDX2 (c, d) expression by immunohistochemistry. a, b This patient (Supplementary Table 1, Case 2) had a stage II (pT4a N0) adenocarcinoma of the cecum (a, H&E, ×100) with isolated loss of PMS2 expression and negative SATB2 expression within the tumor (b, ×100). Positive SATB2 expression is seen within adjacent non-neoplastic colonic crypt epithelium (b, right half of image). Positive CDX2 expression was identified within the adenocarcinoma (not shown). This patient did not receive adjuvant chemotherapy, developed tumor recurrence 7 months after surgical resection, and died of disease 17 months after initial presentation. c, d This patient (Supplementary Table 1, Case 1) had a stage II (pT4a N0) adenocarcinoma of the sigmoid colon (c, H&E, ×100) with loss of MSH2 and MSH6 expression and negative CDX2 expression within the tumor (d, ×100). Positive expression is seen within adjacent non-neoplastic colonic crypt epithelium (d, top of image). SATB2-positive expression was identified within the adenocarcinoma (not shown). This patient received adjuvant FOLFOX chemotherapy but developed liver metastasis 12 months after surgical resection and died of disease 22 months after initial presentation
Of the 514 colonic adenocarcinomas, 506 had adequate tumor tissue available to evaluate CDX2 immunohistochemical expression while histologic review was performed in 362 (72%) tumors. Fifty-five of 506 tumors (11%) had negative CDX2 immunohistochemistry expression (Fig. 1). Similar to SATB2, negative CDX2 expression was also associated with adverse histologic features (Table 1). Compared with CDX2-positive tumors, CDX2-negative tumors more often exhibited high tumor grade (50% vs. 12%, p < 0.001), lymphatic invasion (83% vs. 51%, p = 0.002), venous invasion (42% vs 23%, p = 0.04), and perineural invasion (42% vs. 22%, p = 0.03). However, neither mucinous or signet ring cell differentiation was more frequently observed in CDX2-negative tumors (both with p > 0.05). In addition, CDX2-negative tumors more frequently displayed tumor infiltrating lymphocytes (46% vs. 17%, p < 0.001). In contrast, there was no difference in tumor infiltrating lymphocytes within tumors stratified by SATB2 expression.
SATB2 and CDX2 expression patterns are associated with BRAF V600E mutation and mismatch repair protein deficiency
SATB2 immunohistochemistry expression
SATB2 expression was significantly affected in adenocarcinomas with a BRAF V600E mutation and mismatch repair protein deficiency (Table 2). Negative SATB2 expression was more often identified in adenocarcinomas with a BRAF V600E mutation compared with BRAF wild-type tumors (22% vs. 9%, p = 0.002) and the mean SATB2 H-score was significantly lower in tumors with a BRAF V600E mutation (118 vs. 154, p < 0.001). There was no difference in the proportion of adenocarcinomas with negative SATB2 expression stratified by mismatch repair protein status (mismatch repair protein deficient, 15% vs. mismatch repair protein proficient, 13%, p = 0.5). However, mismatch repair protein deficient adenocarcinomas did display a significantly lower mean SATB2 H-score compared with mismatch repair protein proficient adenocarcinomas (119 vs. 148, p < 0.001). SATB2 expression was not affected by KRAS exon 2 or exon 3 mutation status.
Although loss of SATB2 expression was significantly associated with BRAF V600E mutation, the significantly high proportion of mucinous (50%) and signet ring cell (16%) differentiation observed in SATB2-negative colonic adenocarcinomas were not associated with the BRAF V600E mutation. In particular, of the 101 tumors with mucinous differentiation (Table 1), 66 were BRAF wild-type and 35 had a BRAF V600E mutation. Twenty-two of 66 (33%) BRAF wild-type tumors and 10 of 35 (29%) BRAF-mutated tumors demonstrated negative SATB2 expression (p = 0.3). Among the tumors with signet ring cell differentiation, 11 of the 15 tumors were BRAF wild-type while the remaining 4 had a BRAF V600E mutation. Within this group, 5 (45%) BRAF wild-type tumors and 2 (50%) BRAF-mutated tumors demonstrated negative SATB2 expression (p = 1.0).
CDX2 immunohistochemistry expression
In contrast to SATB2, negative CDX2 expression was significantly more often observed in mismatch repair protein deficient adenocarcinomas compared with mismatch repair protein proficient adenocarcinomas (26% vs. 5%, p < 0.001) (Table 2). In addition, mismatch repair protein deficient tumors had significantly lower mean CDX2 H-score (80 vs. 163, p < 0.001). Similar to SATB2, CDX2 expression was also significantly associated with BRAF V600E mutation (BRAF V600E 23% vs. BRAF wild-type 5%, p < 0.001; mean H-score: 76 vs. 161, p < 0.001). The proportion of KRAS exon 2 or exon 3 mutated tumors that demonstrated negative CDX2 expression was significantly less than that of KRAS wild-type tumors (3% vs. 10%, p = 0.04). However, the mean CDX2 H-scores were similar between the two groups.
SATB2 and CDX2 mRNA expression in the Cancer Genome Atlas colon cancer dataset
To further examine SATB2 and CDX2 expression in colonic adenocarcinomas, we also queried the relationships between SATB2 and CDX2 mRNA expression, status of microsatellite instability, BRAF V600E mutation, and KRAS exon 2 or exon 3 mutations in the colonic adenocarcinoma dataset in the Cancer Genome Pan Cancer Atlas [37,38,39]. Similar to the immunohistochemistry expression patterns we observed, mRNA expression levels of both SATB2 and CDX2 were significantly decreased in high-level microsatellite instability colonic adenocarcinomas and tumors with BRAF V600E mutation compared with microsatellite stable adenocarcinomas or BRAF V600E wild-type tumors (p < 0.0001 for all) (Fig. 2). SATB2 and CDX2 mRNA expression levels were not associated with KRAS exons 2 and/or 3 mutations (p > 0.05 for both).
Combined SATB2 and CDX2 immunohistochemistry expression
The combined SATB2 and CDX2 immunohistochemistry expression profile was also affected by mismatch repair protein and BRAF mutation status (Table 3). Most colonic adenocarcinomas demonstrated concurrent positive SATB2 and CDX2 expression (SATB2+/CDX2+). Concurrent negative SATB2 and CDX2 expression (SATB2−/CDX2−) was only rarely observed and was identified in only 6% of mismatch repair protein deficient and 6% of BRAF V600E mutant colonic adenocarcinomas. In contrast, loss of either SATB2 or CDX2 (SATB2+/CDX2– or SATB2−/CDX2+) was identified in 27% of mismatch repair protein deficient tumors and 30% of BRAF V600E mutant tumors, respectively. Taken together, 33% of mismatch repair deficient colonic adenocarcinomas and 36% of BRAF V600E mutant adenocarcinomas demonstrated negative SATB2 expression and/or negative CDX2 expression (SATB2+/CDX2–, SATB2−/CDX2+, or SATB2−/CDX2−). Furthermore, mismatch repair protein deficient tumors more often demonstrated negative expression of CDX2 and/or SATB2 compared with mismatch repair proficient tumors (33% vs. 15%, p < 0.001). A similar association was also observed in tumors with BRAF V600E mutation (36% vs. 13%, p < 0.001).
SATB2 and CDX2 immunohistochemistry expression patterns are associated with poor prognosis in mismatch repair protein deficient colonic adenocarcinoma
A total of 429 (84%) patients with both known SATB2 and CDX2 expression status had clinical follow-up data for disease-specific survival analysis, including 131 of 146 (90%) patients with mismatch repair protein deficient adenocarcinomas and 298 of 353 (84%) patients with mismatch repair protein proficient adenocarcinomas. The median follow-up interval was 32 months (range: 1 to 100 months) from the time of the initial diagnosis. There were 91 deaths from disease occurring between 2 and 56 months from the time of diagnosis. Clinical follow-up data for disease-free survival were only available for the University of Pittsburgh Medical Center cohort. A total of 221 (43%) patients with both known SATB2 and CDX2 expression status had clinical follow-up data for disease-free survival analysis, including 93 patients with mismatch repair protein deficient adenocarcinomas and 128 patients with mismatch repair protein proficient adenocarcinomas. For patients with mismatch repair protein proficient tumors, neither negative CDX2 nor negative SATB2 had significant association with disease-specific survival or disease-free survival (p > 0.05 for all).
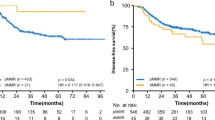
For patients with mismatch repair protein deficient adenocarcinomas, negative SATB2 expression alone (Fig. 3a) and negative CDX2 expression alone (Fig. 3b) within the tumor were significantly associated with decreased disease-specific survival compared with positive expression using Kaplan–Meier survival functions (p = 0.001 for both). Furthermore, in these patients, when SATB2 expression and CDX2 expression status were combined, negative expression of CDX2 and/or SATB2 (SATB2+/CDX2−, SATB2−/CDX2+, and SATB2−/CDX2−) was also significantly associated with decreased disease-specific survival compared with positive expression of both proteins (SATB2+/CDX2+) (Fig. 3c).
a Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient colon cancer of all stages stratified by SATB2 expression. Patients with SATB2-negative tumors had reduced disease-specific survival compared with patients with SATB2-positive tumors (p = 0.001). b Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient colon cancer of all stages stratified by CDX2 expression. Patients with CDX2-negative tumors had reduced disease-specific survival compared with patients with CDX2-positive tumors (p = 0.001). c Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient colon cancer of all stages stratified by SATB2/CDX2 expression. Patients with SATB2 and/or CDX2-negative tumors had reduced disease-specific survival compared with patients with CDX2-positive/SATB2-positive tumors (p < 0.001). d Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient colon cancer with stage II or III disease stratified by SATB2/CDX2 expression. Patients with SATB2 and/or CDX2-negative tumors had reduced disease-specific survival compared with patients with CDX2-positive/SATB2-positive tumors (p < 0.001). e Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient SATB2 and/or CDX2-negative colon cancer of all stages with patients with mismatch repair protein proficient colon cancer of all stages. Patients with mismatch repair protein deficient SATB2 and/or CDX2-negative tumors had similar disease-specific survival compared with patients with mismatch repair protein proficient tumors (p = 0.7). f Kaplan–Meier survival curve comparing the disease-specific survival of patients with mismatch repair protein deficient SATB2 and/or CDX2-negative colon cancer with stage II or III disease with patients with mismatch repair protein proficient colon cancer with stage II or III disease. Patients with mismatch repair protein deficient SATB2 and/or CDX2-negative tumors had similar disease-specific survival compared with patients with mismatch repair protein proficient tumors (p = 0.5) in the stage II and stage III setting
Since the number of patients at risk was the largest when combined SATB2 and CDX2 expression was considered, the combined expression was used in the subsequent multivariable analysis. Using Cox proportional hazards modeling for the cohort of patients with mismatch repair protein deficient tumors (Table 4), features associated with decreased disease-specific survival on both univariate and multivariate analysis were stage and the presence of either negative SATB2 or negative CDX2 expression (multivariate hazard ratio 24.3, 2.2–56.6, 95% confidence interval; p = 0.009). The presence of venous invasion, lymphatic invasion, and perineural invasion were associated with decreased disease-specific survival on univariate analysis (all with p < 0.05) but not on multivariate analysis.
When limited to patients with stage II or III mismatch repair protein deficient tumors (n = 99), negative expression of CDX2 and/or SATB2 was significantly associated with decreased disease-specific survival compared with positive expression of both proteins (SATB2+/CDX2+) (p < 0.001) (Fig. 3d). Of the 99 patients with stage II or III mismatch repair protein deficient tumors, seven patients died of disease (three with stage II disease and four with stage III disease) (Supplementary Table 1). All seven patients had tumors with either negative SATB2 expression (n = 2) or negative CDX2 expression (n = 5). None of the patients with stage II or III mismatch repair protein deficient tumors with dual SATB2+/CDX2+ expression developed recurrence or died of disease.
The effect of negative CDX2 and/or SATB2 expression on disease-specific survival in mismatch repair protein deficient colonic adenocarcinomas was tentatively investigated by comparing disease-specific survival of patients with mismatch repair protein deficient tumors with CDX2 and/or SABT2 negative expression with that of patients with mismatch repair protein proficient tumors regardless of CDX2 or SATB2 expression status in our cohort. Patients with negative expression of CDX2 and/or SATB2 in their mismatch repair protein deficient adenocarcinomas had a similar disease-specific survival to patients with mismatch repair protein proficient adenocarcinomas in all stage groups (Fig. 3e) and in the stage II and III setting (Figure 3f) (p = 0.7 and 0.5, respectively). These findings might indicate that negative expression of CDX2 and/or SATB2 in mismatch repair deficient colonic adenocarcinoma has a negative impact on patient survival that could offset the prognostic benefit conferred by the mismatch repair protein deficiency.
Similar associations were also observed for disease-free survival of all patients with mismatch repair protein deficient adenocarcinomas and patients with stage II or III mismatch repair protein deficient adenocarcinomas. In particular, negative expression of CDX2 and/or SATB2 was significantly associated with decreased disease-free survival compared with positive expression of both proteins for all patients with mismatch repair deficient adenocarcinomas (Fig. 4a) and in the subgroup of patients with stage II or III adenocarcinomas (Fig. 4b).
a Kaplan–Meier survival curve comparing the disease-free survival of patients with mismatch repair protein deficient colon cancer of all stages stratified by SATB2/CDX2 expression. Patients with SATB2 and/or CDX2-negative tumors had reduced disease-free survival compared with patients with CDX2-positive/SATB2-positive tumors (p < 0.001). b Kaplan–Meier survival curve comparing the disease-free survival of patients with stage II or stage III mismatch repair protein deficient tumors stratified by SATB2/CDX2 expression. Patients with SATB2 and/or CDX2-negative tumors had reduced disease-free survival compared with patients with CDX2-positive/SATB2-positive tumors (p < 0.001)
Discussion
The primary motivation for this study was to evaluate the prognostic significance of SATB2 and CDX2 expression in colonic adenocarcinoma in a cohort enriched for patients with mismatch repair protein deficient tumors. Our results indicate that negative immunohistochemical expression of SATB2 and/or CDX2 is associated with reduced disease-specific survival in patients with mismatch repair protein deficient tumors. Death from disease in patients with stage II or III mismatch repair protein deficient tumors only occurred in patients with negative expression of either SATB2 or CDX2 in their tumors, indicating that SATB2 and CDX2 are potentially important prognostic biomarkers in this setting. SATB2 and CDX2 expression patterns did not affect patient survival in patients with colonic adenocarcinomas that were mismatch repair protein proficient or had BRAF or KRAS mutations. Our results suggest that SATB2 and CDX2 may be helpful prognostic biomarkers, particularly in patients with mismatch repair protein deficient colon cancer.
Although tumor stage remains the most important prognostic variable in colon cancer, ~20% of patients with stage II colon cancer will develop distant metastasis and die from metastatic disease [2]. Currently, decision-making on the use of adjuvant chemotherapy in patients with stage II disease is based on a number of histopathologic factors, including mismatch repair protein expression within the tumor [40]. Mismatch repair protein deficient tumors are more often identified in patients with stage II disease (~20%) compared with stage III (9–12%) or stage IV disease (3–4%) [5, 12, 41,42,43]. In patients with stage II disease, mismatch repair protein deficiency has generally been associated with improved survival [7,8,9,10,11,12]; however, not all studies have verified the association between mismatch repair protein deficiency and improved survival in patients with stage II disease [43, 44]. In a recent meta-analysis of 19 studies including 5998 patients with stage II colon cancer, Gkekas et al. identified no significant overall or disease-free survival benefit in patients with mismatch repair protein deficient tumors [43]. Similarly, in patients with stage III colon cancer, the prognostic impact of mismatch repair protein deficiency remains controversial with most studies indicating improved survival [4, 5] while other studies identifying reduced disease-specific survival in patients with mismatch repair protein deficient stage III colon cancer [12]. These conflicting literature reports suggest that mismatch repair protein deficient colon cancers are clinically heterogeneous, and identification of biomarkers that can predict clinically aggressive disease in mismatch repair protein deficient tumors may be helpful. In our analysis, negative SATB2 expression and/or negative CDX2 expression is identified in more than one-third of mismatch repair protein deficient colon cancers and is associated with reduced disease-specific survival. Our data suggest that negative expression of SATB2 and/or CDX2 identifies patients with clinically aggressive mismatch repair protein deficient colon cancer, including in the stage II or III setting.
Very limited literature data exist on the prognostic significance of SATB2 and CDX2 expression in colon cancer, particularly in patients with mismatch repair protein deficient tumors (Table 5). To our knowledge, no study has concurrently evaluated the prognostic significance of both markers. Using a tissue microarray approach, Eberhardt et al. analyzed 527 colorectal carcinomas and identified negative SATB2 expression in 152 (28.8%) of tumors [26]. In their study, negative SATB2 expression was frequently identified in patients with mismatch repair protein deficient tumors (42/77, 55%), a much higher proportion than the 15% negative SATB2 expression in mismatch repair protein deficient tumors identified in our analysis. While negative SATB2 expression was an independent predictor of decreased disease-specific survival in colon cancer, Eberhardt et al. did not identify any prognostic effect of SATB2 expression in patients with mismatch repair protein deficient tumors. There are a number of differences between our study and Eberhardt et al. that may help to explain the different results. First, our cohort was enriched for patients with mismatch repair protein deficient colon cancer (146 patients) compared with Eberhardt et al. (77 patients). Second, different SATB2 antibody clones were used in each study that may explain the vastly different proportion of negative SATB2 mismatch repair protein deficient tumors (55% vs. 15%). Finally, Eberhardt et al. stratified SATB2 expression into negative, intermediate, and high expression, and it is unclear if they specifically evaluated the effect of negative SATB2 expression on survival for patients with mismatch repair protein deficient tumors. Wang et al. evaluated 146 colorectal carcinomas and demonstrated that “low level” SATB2 expression (defined as either no staining or weak expression in < 25% of tumor cells) was independently associated with poor overall survival [25]. However, Wang et al. did not specifically evaluate the impact of negative SATB2 expression on patient survival in patients with mismatch repair protein deficient tumors.
Previous literature reports have demonstrated that CDX2 is a prognostic biomarker in colon cancer [13, 14, 27, 45,46,47,48], although not all studies have confirmed the prognostic effect of CDX2 expression [49,50,51]. Over the last decade, negative CDX2 expression in mismatch repair protein deficient colorectal carcinoma has been well-documented in the literature [27,28,29,30]. However, very few literature reports have specifically evaluated the prognostic significance of CDX2 expression in mismatch repair protein deficient colon cancer (Table 5). In our series, negative CDX2 expression was identified in 29% of mismatch repair protein deficient tumors and was associated with reduced disease-specific survival. Similarly, Kim et al. identified reduced disease-specific survival for patients with high-level microsatellite instability tumors and negative CDX2 expression, although this was not significant on multivariable analysis or in an analysis limited to patients with low-stage (I–II) disease [27]. Bruun et al. evaluated 164 patients with mismatch repair protein deficient colorectal carcinomas and did not identify an effect of negative CDX2 immunohistochemistry expression on overall survival [46]. Using CDX2 mRNA expression in a cohort of stage II/III colon cancer patients, Pilati et al. did not identify negative CDX2 expression as a predictor of overall survival in mismatch repair protein deficient tumors [45]. The reason for the different results between our analysis and previous literature reports is not entirely clear. Our analysis and that of Kim et al. used disease-specific survival as study endpoints; however, Bruun et al. and Pilati et al. used overall survival as an endpoint. Overall survival is a reasonable endpoint only if the observed mortality can largely be attributed to cancer rather than other underlying medical conditions. However, differences in disease-specific and overall survival can be substantial in patients with mismatch repair protein deficient tumors as these patients more often present with low-stage disease and often occur in older individuals with significant underlying medical conditions that could lead to death from a cause unrelated to cancer. Thus, for patients with mismatch repair protein deficient tumors, disease-specific survival is likely a more accurate predictor of outcome than overall survival. Additional large, well-characterized patient populations with mismatch repair protein deficient colon cancer and with extended follow-up are needed to further determine the prognostic impact of SATB2 and CDX2 expression.
In a recent study, we identified SATB2 as a relatively specific marker of colorectal carcinoma and that in combination with CDX2, SATB2+/CDX2+ expression has a 94% specificity for lower gastrointestinal tract adenocarcinoma [18]. The proportion of mismatch repair protein deficient colon cancers with negative SATB2 expression in our current study (16%) is lower than in our previous analysis (31%), although the number of mismatch repair protein deficient colon cancers analyzed is much larger in our current analysis (146 tumors versus 42 tumors in our previous study) [18]. In our current study, mismatch repair protein deficient tumors did display a significantly lower mean SATB2 H-score compared with mismatch repair protein proficient tumors, and this finding is supported by the Cancer Genome Atlas dataset that also identified significantly reduced SATB2 mRNA expression in high-level microsatellite instability colon cancer compared with microsatellite stable colon cancer. The reason for reduced SATB2 protein and mRNA expression observed in mismatch repair protein deficient colon carcinoma is unknown. A recent study on ARID1A, another transcription factor functioning in chromatin remodeling as part of the SWI/SNF complex and a tumor suppressor, showed that ARID1A recruited DNA mismatch repair protein MSH2 to chromatin during DNA replication and thus protected DNA from damage [52]. Loss of ARID1A expression or ARID1A inactivation, in contrast, compromised repair of DNA damage and was associated with microsatellite instability [52]. Since SATB2 also functions in chromatin remodeling [16] albeit belonging to a protein family different from ARID1A, its interactions with mismatch repair proteins may be worth future investigation. On the other hand, reduced CDX2 expression in mismatch repair protein deficient colorectal carcinoma has been hypothesized to occur as a result of passenger mutations within simple repeat sequences in the CDX2 gene [53, 54] or epigenetic silencing by CpG island promoter hypermethylation [27, 30, 47, 48, 55, 56]. It is likely that alternative mechanisms lead to reduced SATB2 expression in mismatch repair protein deficient tumors as only 6% of mismatch repair protein deficient colonic adenocarcinomas demonstrated concurrent negative CDX2 and SATB2 expression.
Our analysis has limitations including the retrospective design, the lack of rigorously standardized treatment in retrospective analyses, and the inherent issue of heterogeneity of protein expression within tumors within a tissue microarray approach. We attempted to address the issue of heterogeneity by performing whole tissue section immunohistochemistry in all available mismatch repair protein deficient tumors that demonstrated negative SATB2 and/or CDX2 expression on the tissue microarray analysis. The whole tissue section immunohistochemistry confirmed the tissue microarray analysis in 34/37 (92%) of cases. Only three patients (8%) had tumors with weak and heterogenous CDX2 expression on whole tumor sections that was not observed in the tissue cores selected for the tissue microarray analysis. These data suggest that the tissue microarray approach adequately reflects SATB2 and CDX2 expression patterns within mismatch repair protein deficient tumors. In addition, although the number of cases is relatively small by some standards, our analysis is the largest study to date correlating SATB2 and CDX2 expression in mismatch repair protein deficient tumors with patient survival. Our study represents a series of colon cancers resected at large academic medical centers with its inherent referral bias. Given the retrospective design, we were also not able to control for chemotherapy treatment, including anti-PD-1 therapy. While many patients with mismatch repair protein deficient tumors demonstrate responsiveness to anti-PD-1 therapy, not all patients with mismatch repair protein deficient colonic adenocarcinoma respond to this treatment [57, 58]. Histopathologic and molecular factors underlying the lack of response to anti-PD-1 therapy in patients with mismatch repair protein deficient colonic adenocarcinoma are unclear, and additional study, including evaluating the effect of SATB2 and CDX2 expression on treatment response, is needed.
In conclusion, SATB2 and CDX2-negative immunohistochemical expression is associated with reduced disease-specific and disease-free survival in patients with mismatch repair protein deficient colonic adenocarcinomas, including in the stage II–III setting, but not in patients with mismatch repair protein proficient tumors. Our results indicate that SATB2 and CDX2 immunohistochemistry can help identify patients with mismatch repair protein deficient tumors that may benefit from a more aggressive adjuvant therapeutic approach following surgery. Given the retrospective design of our study, these results will need to be validated prospectively using a larger cohort of patients with mismatch repair protein deficient colonic adenocarcinomas to confirm the prognostic value of SATB2 and CDX2 immunohistochemistry.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Bockelman C, Engelmann BE, Kaprio T, Hansen TF, Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54:5–16.
Kannarkatt J, Joseph J, Kurniali PC, Al-Janadi A, Hrinczenko B. Adjuvant chemotherapy for stage II colon cancer: a Clinical Dilemma. J Oncol Pract. 2017;13:233–41.
Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87.
Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99.
Landau MA, Zhu B, Akwuole FN, Pai RK. Site-specific differences in colonic adenocarcinoma: KRAS mutations and high tumor budding are more frequent in cecal adenocarcinoma. Am J Surg Pathol. 2018;42:351–8.
Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18.
Klingbiel D, Saridaki Z, Roth AD, Bosman FT, Delorenzi M, Tejpar S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann Oncol. 2015;26:126–32.
Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57.
Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–75.
Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–226.
Mohan HM, Ryan E, Balasubramanian I, Kennelly R, Geraghty R, Sclafani F, et al. Microsatellite instability is associated with reduced disease specific survival in stage III colon cancer. Eur J Surg Oncol. 2016;42:1680–6.
Zhang BY, Jones JC, Briggler AM, Hubbard JM, Kipp BR, Sargent DJ, et al. Lack of caudal-type homeobox transcription factor 2 expression as a prognostic biomarker in metastatic colorectal cancer. Clin Colorectal Cancer. 2017;16:124–8.
Dalerba P, Sahoo D, Paik S, Guo X, Yothers G, Song N, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374:211–22.
Bae JM, Lee TH, Cho NY, Kim TY, Kang GH. Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2015;21:1457–67.
FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–501.
Magnusson K, de Wit M, Brennan DJ, Johnson LB, McGee SF, Lundberg E, et al. SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol. 2011;35:937–48.
Ma C, Olevian DC, Lowenthal BM, Jayachandran P, Kozak MM, Chang DT, et al. Loss of SATB2 expression in colorectal carcinoma Is associated with DNA mismatch repair protein deficiency and BRAF mutation. Am J Surg Pathol. 2018;42:1409–17.
Lin F, Shi J, Zhu S, Chen Z, Li A, Chen T, et al. Cadherin-17 and SATB2 are sensitive and specific immunomarkers for medullary carcinoma of the large intestine. Arch Pathol Lab Med. 2014;138:1015–26.
Dragomir A, de Wit M, Johansson C, Uhlen M, Ponten F. The role of SATB2 as a diagnostic marker for tumors of colorectal origin: results of a pathology-based clinical prospective study. Am J Surg Pathol. 2014;141:630–8.
Moh M, Krings G, Ates D, Aysal A, Kim GE, Rabban JT. SATB2 expression distinguishes ovarian metastases of colorectal and appendiceal origin from primary ovarian tumors of mucinous or endometrioid type. Am J Surg Pathol. 2016;40:419–32.
Strickland S, Wasserman JK, Giassi A, Djordjevic B, Parra-Herran C. Immunohistochemistry in the diagnosis of mucinous neoplasms involving the ovary: the added value of SATB2 and biomarker discovery through protein expression database mining. Int J Gynecol Pathol. 2016;35:191–208.
Li Z, Roth R, Rock JB, Lehman A, Marsh WL, Suarez A, et al. Dual immunostain with SATB2 and CK20 differentiates appendiceal mucinous neoplasms from ovarian mucinous neoplasms. Am J Clin Pathol. 2017;147:484–91.
Mansour MA, Hyodo T, Ito S, Kurita K, Kokuryo T, Uehara K, et al. SATB2 suppresses the progression of colorectal cancer cells via inactivation of MEK5/ERK5 signaling. FEBS J. 2015;282:1394–405.
Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM, et al. Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol. 2009;219:114–22.
Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlen M, Ericson Lindquist K, et al. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931–8.
Kim JH, Rhee YY, Bae JM, Cho NY, Kang GH. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am J Surg Pathol. 2013;37:1532–41.
Lugli A, Tzankov A, Zlobec I, Terracciano LM. Differential diagnostic and functional role of the multi-marker phenotype CDX2/CK20/CK7 in colorectal cancer stratified by mismatch repair status. Mod Pathol. 2008;21:1403–12.
McGregor DK, Wu TT, Rashid A, Luthra R, Hamilton SR. Reduced expression of cytokeratin 20 in colorectal carcinomas with high levels of microsatellite instability. Am J Surg Pathol. 2004;28:712–8.
Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–43.
Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB, et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol. 2002;161:1557–65.
Greenson JK, Bonner JD, Ben-Yzhak O, Cohen HI, Miselevich I, Resnick MB, et al. Phenotype of microsatellite unstable colorectal carcinomas: well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am J Surg Pathol. 2003;27:563–70.
Lugli A, Kirsch R, Ajioka Y, Bosman F, Cathomas G, Dawson H, et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30:1299–311.
Kakar S, Shi C, Berho M, Driman DK, Fitzgibbons P, Frankel WL, et al. Protocol for the examination of specimens from patients with primary carcinoma of the colon and rectum. In: CAP (College of American Pathologists) Cancer Protocols, 2018 [cited 1 November 2018]. https://cap.objects.frb.io/protocols/cp-gilower-colonrectum-17protocol-4010.pdf
Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744–52.
Yousem SA, Nikiforova M, Nikiforov Y. The histopathology of BRAF-V600E-mutated lung adenocarcinoma. Am J Surg Pathol. 2008;32:1317–21.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–12.
Benson AB, 3rd, Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:370–98.
Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–73.
Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74.
Gkekas I, Novotny J, Pecen L, Strigard K, Palmqvist R, Gunnarsson U. Microsatellite instability as a prognostic factor in stage II colon cancer patients, a meta-analysis of published literature. Anticancer Res. 2017;37:6563–74.
Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–72.
Pilati C, Taieb J, Balogoun R, Marisa L, de Reynies A, Laurent-Puig P. CDX2 prognostic value in stage II/III resected colon cancer is related to CMS classification. Ann Oncol. 2017;28:1032–5.
Bruun J, Sveen A, Barros R, Eide PW, Eilertsen I, Kolberg M, et al. Prognostic, predictive, and pharmacogenomic assessments of CDX2 refine stratification of colorectal cancer. Mol Oncol. 2018;12:1639–55.
Graule J, Uth K, Fischer E, Centeno I, Galvan JA, Eichmann M, et al. CDX2 in colorectal cancer is an independent prognostic factor and regulated by promoter methylation and histone deacetylation in tumors of the serrated pathway. Clin Epigenetics. 2018;10:120.
Baba Y, Nosho K, Shima K, Freed E, Irahara N, Philips J, et al. Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res. 2009;15:4665–73.
Olsen J, Eiholm S, Kirkeby LT, Espersen ML, Jess P, Gogenur I, et al. CDX2 downregulation is associated with poor differentiation and MMR deficiency in colon cancer. Exp Mol Pathol. 2016;100:59–66.
Coebergh van den Braak RR, Martens JW, Ijzermans JN. CDX2 as a prognostic biomarker in colon cancer. N Engl J Med. 2016;374:2182.
Schirripa M, Loupakis F, Lenz HJ. CDX2 as a prognostic biomarker in colon cancer. N Engl J Med. 2016;374:2183.
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62.
Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S, et al. CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proc Natl Acad Sci USA. 2012;109:E3196–3205.
Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8.
Walsh MD, Clendenning M, Williamson E, Pearson SA, Walters RJ, Nagler B, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–56.
Olsen J, Espersen ML, Jess P, Kirkeby LT, Troelsen JT. The clinical perspectives of CDX2 expression in colorectal cancer: a qualitative systematic review. Surg Oncol. 2014;23:167–76.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch repair deficiency. N Engl J Med. 2015;372:2509–20.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ma, C., Olevian, D., Miller, C. et al. SATB2 and CDX2 are prognostic biomarkers in DNA mismatch repair protein deficient colon cancer. Mod Pathol 32, 1217–1231 (2019). https://doi.org/10.1038/s41379-019-0265-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0265-1
This article is cited by
-
Loss of SATB2 and CDX2 expression is associated with DNA mismatch repair protein deficiency and BRAF mutation in colorectal cancer
Medical Molecular Morphology (2024)
-
Delineating the intra-patient heterogeneity of molecular alterations in treatment-naïve colorectal cancer with peritoneal carcinomatosis
Modern Pathology (2022)
-
Comprehensive clinicopathologic, molecular, and immunologic characterization of colorectal carcinomas with loss of three intestinal markers, CDX2, SATB2, and KRT20
Virchows Archiv (2022)
-
Intratumoral budding and automated CD8-positive T-cell density in pretreatment biopsies can predict response to neoadjuvant therapy in rectal adenocarcinoma
Modern Pathology (2021)
-
Loss of CDX2 in colorectal cancer is associated with histopathologic subtypes and microsatellite instability but is prognostically inferior to hematoxylin–eosin-based morphologic parameters from the WHO classification
British Journal of Cancer (2021)