Abstract
Colorectal neuroendocrine carcinomas, both small cell and large cell types, are highly aggressive tumors with poor prognosis compared with colorectal adenocarcinoma. The molecular drivers of neuroendocrine carcinoma are best defined in small cell lung cancer, which shows near-universal genomic alterations in TP53 and RB1. The genetics of colorectal neuroendocrine carcinoma remain poorly understood; recent studies demonstrated infrequent RB1 alterations and genetics closely resembling colorectal adenocarcinoma. To better define the molecular pathogenesis of colorectal neuroendocrine carcinoma, we performed capture-based next-generation sequencing on 25 cases and evaluated for expression of p53, Rb, p16, and high-risk human papillomavirus (HR-HPV) subtypes using immunohistochemistry, in situ hybridization, and polymerase chain reaction. Rb/E2F pathway dysregulation was identified in nearly all cases (23/25, 92%) and occurred via three distinct mechanisms. First, RB1 genomic alteration was present in 56% (14/25) of cases and was accompanied by Rb protein loss, high p16 expression, and absence of HR-HPV; these cases also had frequent genomic alterations in TP53, the PI3K/Ras and Wnt pathways, as well as in DNA repair genes, with 4/14 cases being hypermutated. Second, 16% (4/25) of cases, all left-sided, had TP53 alteration without RB1 alteration; half of these harbored high-level amplifications in CCNE1 and MYC or MYCN and arose in patients with ulcerative colitis. Finally, 28% (7/25) of cases, all rectal or anal, lacked genomic alterations in RB1 or TP53 but were positive for HR-HPV. Our data demonstrate that Rb/E2F pathway dysregulation is essential in the pathogenesis of colorectal neuroendocrine carcinoma, akin to neuroendocrine carcinomas in other anatomic sites. Moreover, colorectal neuroendocrine carcinomas stratify into three distinct molecular subgroups, which can be differentiated based on Rb protein and HR-HPV status. HR-HPV infection represents a distinct mechanism for Rb and p53 inactivation in cases lacking genomic alterations in either gene. Differential treatment strategies for hypermutated and HPV-driven cases could improve patient outcomes.
Similar content being viewed by others
Introduction
Neuroendocrine neoplasms occur in multiple anatomic sites and are divided into two major categories: well-differentiated neuroendocrine tumors (also called carcinoid in the lung) and poorly differentiated neuroendocrine carcinomas [1]. Neuroendocrine tumors and neuroendocrine carcinomas were once considered to comprise a single biological spectrum, as both produce neurosecretory proteins (e.g., synaptophysin and chromogranin); however, recent clinicopathologic and genome-wide sequencing studies [2,3,4] support the concept that neuroendocrine carcinoma largely represents a distinct entity that harbors different genomic alterations. This conclusion is supported by observations that neuroendocrine carcinomas rarely evolve from or coexist with neuroendocrine tumors but rather are often admixed with adenocarcinoma or squamous cell carcinoma [4, 5]. Alternatively, neuroendocrine carcinomas also emerge in the setting of relapse as a mechanism of therapeutic resistance, including from castration-resistant prostate cancer treated with androgen deprivation therapy [6] and EGFR-mutant lung cancer treated with tyrosine kinase inhibitors [7]. Thus, neuroendocrine carcinoma can arise de novo or via progression from an epithelial neoplasm, including as a mechanism of treatment resistance [8].
Regardless of anatomic site, neuroendocrine carcinomas are high-grade neoplasms that grow as sheets, ribbons, or nests, with frequent necrosis, high mitotic activity, and high Ki-67 proliferative indexes (often >70%). Neuroendocrine carcinomas are further subdivided into small cell and large cell subtypes, although many neuroendocrine carcinomas show both small cell and large cell areas. Patients frequently have metastatic disease at the time of diagnosis and carry a poor prognosis, with a median survival of 7.6 months for lung neuroendocrine carcinoma, 7.5 months for gastrointestinal neuroendocrine carcinoma, and 2.5 months for neuroendocrine carcinoma of unknown primary [9].
The genetics of neuroendocrine carcinoma have been best described in the lung, pancreas, and prostate, which have revealed that approximately 80% of neuroendocrine carcinomas show dual alterations in the p53-encoding gene TP53 and the retinoblastoma (Rb)-encoding gene RB1, despite having highly variable mutation rates [8]. A comprehensive whole-genome sequencing study of 110 small cell lung carcinomas, the prototypical neuroendocrine carcinoma, found near-universal biallelic inactivation of TP53 and RB1 via various genomic alteration types, including mutations, deletions, and complex rearrangements, particularly in the case of RB1 [3]. This study established TP53 and RB1 as obligatory tumor suppressors in the pathogenesis of small cell lung cancer and suggested that targeted gene panels used in other studies may not fully capture all RB1-aberrant tumors. In contrast, TP53 and RB1 are altered in roughly 40 and 5%, respectively, of adenocarcinomas from the same anatomic sites [8] and are infrequently altered in neuroendocrine tumors, which instead harbor frequent mutations in MEN1 (lung, pancreas) and DAXX/ATRX (pancreas) [4, 10, 11].
The genetics of neuroendocrine carcinomas originating at other anatomic sites are less well established. Limited studies have demonstrated an association of neuroendocrine carcinoma with human papillomavirus (HPV) infection in the cervix (HPV 18 ≫ HPV 16) [12] and oropharynx (HPV 16 ≫ 18) [13, 14], with a subset of the latter being associated with conventional HPV-related oropharyngeal squamous cell carcinoma. In addition, one study observed high-risk HPV (HR-HPV) by in situ hybridization in anal (5/5 cases) and rectal (9/11 cases) small cell carcinoma, but only two cases (1 anal, 1 rectal) had high viral copy numbers while the remaining cases showed low HPV copy number [15]. Given the essential role for loss of TP53 and RB1 in the pathogenesis of small cell lung cancer, HPV infection might represent an alternative mechanism for p53 and Rb inactivation via the virally encoded oncoproteins E6 and E7, respectively [16]; however, none of the above studies examined the genomic profiles of neuroendocrine carcinomas that were associated with HR-HPV.
Neuroendocrine carcinomas in the colon or rectum are rare [17], and their molecular pathogenesis remains poorly understood. To date, three studies using either Sanger sequencing or small panel amplicon-based next-generation sequencing examined the molecular features of colorectal neuroendocrine carcinomas and/or mixed adenoneuroendocrine carcinomas [18,19,20]. All three studies observed frequent mutations in BRAF, KRAS, TP53, and APC and established that the neuroendocrine carcinoma component of mixed tumors shares a clonal origin with the glandular component. However, there was contradicting data on the frequency of RB1 dysregulation; the first study showed decreased Rb expression by immunohistochemistry in 56% of neuroendocrine carcinomas [18], whereas the two more recent studies observed infrequent alterations in RB1 [19, 20]. Taken together, the emerging paradigm from prior data is that colorectal neuroendocrine carcinoma more closely resembles colorectal adenocarcinoma and does not require RB1 dysregulation akin to neuroendocrine carcinoma at other sites.
In this study, we sought to define the molecular drivers of colorectal neuroendocrine carcinoma, including the requirement for RB1 inactivation and association with HR-HPV infection, with attention to differences between tumors that arise in the right vs. left colon. We identified 25 cases of colorectal neuroendocrine carcinoma at our institution and evaluated for expression of p53, Rb, and p16 by immunohistochemistry and assessed for HR-HPV infection by in situ hybridization and polymerase chain reaction (PCR), separately analyzing synchronous glandular or squamous components. We additionally profiled genomic alterations in 24/25 cases by capture-based next-generation sequencing of 479 cancer-related genes. Our results demonstrate that Rb/E2F pathway dysregulation is a key feature of colorectal neuroendocrine carcinoma and can be mediated by multiple molecular mechanisms, including HR-HPV infection.
Materials and methods
Case selection
This study was approved by the Institutional Review Board of the University of California, San Francisco. Twenty-five colorectal or anal poorly differentiated neuroendocrine carcinomas were identified in the pathology department archives of the University of California, San Francisco, spanning 1994−2017, for which slides and/or tissue were available. Nine low or intermediate grade (G1/G2) rectal neuroendocrine tumors, spanning years 2006−2015, and 80 colorectal adenocarcinomas (33 right-sided, 6 transverse, 23 descending/sigmoid, and 18 rectal), spanning years 2000−2014, were selected for comparison for immunohistochemical and/or HR-HPV analysis. All specimens were fixed in 10% neutral buffered formalin and embedded in paraffin. All clinical information was obtained from the University of California, San Francisco electronic medical record and from referring institutions. Overall survival was calculated from date of first pathologic diagnosis until date of death and was censored as date of last known alive for patients still alive or lost to follow-up. Survival analyses were performed with STATA 13 Professional software package (STATA/MP 14.2, StataCorp, College Station, TX) using univariate Cox proportional hazards models.
Histopathology
Hematoxylin and eosin-stained sections of each neuroendocrine carcinoma were independently reviewed by five gastrointestinal pathologists (S.K., G.E.K., S.E.U., S.-J.C., N.M.J.) and one lung pathologist (K.J.) to confirm the diagnosis of poorly differentiated neuroendocrine carcinoma and to further classify the tumor as small cell, large cell, or neuroendocrine carcinoma, not otherwise specified (Supplementary Figure S1). The latter category was used for any of the following: (a) the tumor demonstrated heterogeneous areas of small cell and large cell morphology; (b) the tumor was morphologically homogeneous with overlapping features of small cell and large cell; or (c) there were discrepant opinions among the six pathologists. Cases with cytologic and architectural features consistent with well-differentiated neuroendocrine tumors with Ki-67 > 20% (now classified as neuroendocrine tumor Grade 3) were excluded from this study.
Tissue microarray construction
Tissue microarrays were created from 17/25 neuroendocrine carcinomas, 9/9 rectal neuroendocrine tumors, and 80/80 colorectal adenocarcinomas using either 4-mm punch biopsies or multiple 2-mm punch biopsies of formalin-fixed paraffin-embedded tissue. Two punches were used for mixed tumors with spatially distinct neuroendocrine and glandular components.
Immunohistochemistry
Immunohistochemistry was performed on whole tissue sections or tissue microarrays. The following antibodies and dilutions were used: synaptophysin (polyclonal, 1:100, Cell Marque, Rocklin, CA, USA), chromogranin A (LK2H10, 1:4, Cell Marque), p16 (E6H4, prediluted, Ventana Medical Systems, Tucson, AZ, USA), Rb (G3-245, 1:100, BD Biosciences, San Jose, CA, USA), p53 (DO-7, prediluted, Leica Biosystems, Buffalo Grove, IL, USA), p63 (4A4, prediluted, Ventana Medical Systems, Tucson, AZ, USA), p40 (BC28, prediluted, Biocare Medical, Concord, CA, USA), CDX-2 (EP25, prediluted, Leica Biosystems), and Ki-67/MIB-1 (MIB-1, 1:50, DAKO, Carpinteria, CA, USA). Antigen retrieval was as follows: for chromogranin A, p16, p63, and CDX-2, Bond epitope retrieval solution 1 (Leica Biosystems); and for synaptophysin, Rb, p53, p40, and Ki-67/MIB-1, Bond epitope retrieval solution 2 (Leica Biosystems).
For synaptophysin, chromogranin, and CDX-2, positive cytoplasmic (synaptophysin or chromogranin) or nuclear (CDX-2) staining was defined as diffuse (>50%), patchy (10–50%), or focal (<10%) with an intensity of weak, moderate, or strong. For p16, diffuse (>90%) strong nuclear and cytoplasmic staining (i.e. block-positive staining [21]) was considered positive; patchy strong cytoplasmic and nuclear staining was classified as patchy; focal strong or patchy weak staining was considered a wild-type pattern; and lack of staining was considered negative/lost. For Rb, diffuse (>50%) or patchy (10–50%) nuclear staining was considered intact, and lack of staining was considered negative/lost. For p53, diffuse (≥70%) strong homogeneous nuclear staining was considered positive; no nuclear staining was considered negative/lost; and patchy, heterogeneous, weak to moderate nuclear staining was considered a wild-type pattern. All cases showing loss of Rb, p53, and p16 demonstrated internal positive control staining of infiltrating inflammatory cells. Ki-67/MIB-1 staining was scored quantitatively using ImageJ software in the foci with the highest percentage of stained tumor nuclei (at least 500 cells per tumor).
High-risk human papillomavirus in situ hybridization and polymerase chain reaction
Two methods were used to assess HR-HPV status: (1) in situ hybridization, performed at ProPath (Dallas, TX, USA), and (2) PCR. Both methods were performed in 20/25 neuroendocrine carcinomas and 8/9 neuroendocrine tumors; three neuroendocrine carcinomas were evaluated by PCR alone, and two neuroendocrine carcinomas and one neuroendocrine tumor by in situ hybridization alone due to limited tissue availability. All 80 colorectal adenocarcinomas were evaluated by in situ hybridization alone. Positive HR-HPV in situ hybridization was defined as nuclear and cytoplasmic dot-like labeling. A known HPV 16-positive basaloid squamous cell carcinoma of the anus served as a positive control for in situ hybridization and PCR. An RNA in situ hybridization control was performed on each case to ensure RNA integrity.
The protocol for extraction and genotyping of HR-HPV DNA was adapted from published protocols [22]. Briefly, DNA was extracted from formalin-fixed paraffin-embedded material using the WaxFree DNA Extraction Kit. After extraction, DNA concentration was quantitated using a Qubit Fluorometer. Approximately 5 µg of total DNA was diluted in TE buffer to a final volume of 1 mL and subsequently diluted with an additional 1 mL of 100% EtOH for a total volume of 2 mL. The diluted DNA was transferred to a 13 mL round bottom tube and transferred to the Roche Cobas® 4800 system, an automated platform that performs sample preparation and DNA extraction coupled to a thermocycler that performs real-time PCR for HR-HPV and β-globin (Cobas® 4800 HPV Amplification/Detection Kit P/N:05235880190). The system amplifies a sequence of ~200 bp from the L1 region and subsequently uses cleaved fluorescent-labeled specific oligonucleotide probes for detection of 14 HR-HPV subtypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68). The assay also uses β-globin-specific complementary primer pairs in parallel to assess for DNA integrity and potential PCR inhibitors. Samples were classified as negative, invalid (if β-globin Ct value was above the detection limit), or positive for HPV 16, HPV 18, or other HR-HPV subtypes.
Capture-based next-generation DNA sequencing
Sequencing was performed at the University of California, San Francisco Clinical Cancer Genomics Laboratory using an assay that targets the coding regions of 479 cancer-related genes, select introns from 47 genes, and the TERT promoter, with a total sequencing footprint of ~2.9 Mb (UCSF500 panel, Supplementary Table S1). In addition, the assay captures 2000 unique sequences containing common SNPs within regions devoid of constitutional copy number variations, to assist in genome-wide copy number and allelic imbalance analysis. Sequencing libraries were prepared from genomic DNA extracted from tumor and normal paraffin-embedded tissue. Target enrichment was performed by hybrid capture using a custom oligonucleotide library, and sequencing was performed on an Illumina HiSeq 2500. Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using the following software packages: BWA: 0.7.13, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: Appistry v2015.1.1-3.4.46-0ga8e1d99, CNVkit: 0.7.2, Pindel: 0.2.5b8, SATK: Appistry v2015.1.1-1-gea45d62, Annovar: v2016 Feb01, Freebayes: 0.9.20 and Delly: 0.7.2 [23,24,25,26,27,28,29,30,31,32,−33]. Somatic single-nucleotide variants, insertions/deletions, and rearrangements were visualized and verified using Integrated Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and Nexus Copy Number (Biodiscovery, Hawthorne, CA, USA) [28]. Large-scale chromosomal changes were defined as those involving entire chromosomes or chromosome arms. Focal copy number changes were defined as those encompassing less than ten megabases. All reported amplifications in Fig. 4 were focal and had >5 copy (>3.5 fold) increase and all deep deletions were focal and had ≥2 copy loss.
Results
Clinicopathologic features of colorectal neuroendocrine carcinomas
The clinicopathologic features of the 25 colorectal or anal neuroendocrine carcinomas analyzed in this study are summarized in Table 1. Eight cases were in the right colon (5 male, 3 female; mean age 56, range 39–71), one case was in the descending colon (female; age 82), and 16 cases were in the rectum or anus, which showed a male predominance (13 male, 3 female; mean age 49, range 25–72). Among right colon cases, three patients died of disease (median survival 7 months), two patients were alive with disease, and three patients were alive with no evidence of disease, with a minimum follow-up of 11 months. Among left colon cases, 13 patients died of disease (median survival 10 months), one patient was alive with disease, and three patients were alive with no evidence of disease, with a minimum follow-up of 13 months. In 11/25 cases (1 right colon, 10 left), the patient only had a mucosal biopsy and did not undergo surgical resection. For these cases, parameters such as tumor size and lymph node status were assessed based on endoscopic and/or radiologic findings.
All cases were reviewed by an expert panel of six surgical pathologists. The 25 included cases were unanimously classified as poorly differentiated neuroendocrine carcinoma. All neuroendocrine carcinomas had a high Ki-67 proliferation index (mean 65%, range 31–93%). Expression of at least one neuroendocrine marker was required for the diagnosis: all cases demonstrated positive staining for synaptophysin (22 diffuse, 1 patchy, and 2 focal) and 18/25 were positive for chromogranin (13 diffuse, 3 patchy, 2 focal) (Supplemental Table S2, Supplemental Figure S2). Fourteen out of 21 cases showed positive staining for CDX-2 (6 diffuse, 6 patchy, 2 focal), and all cases were negative for the squamous markers p40 and p63 (Supplementary Figure S2, Supplementary Table S2). In 10/25 cases, there was unanimous agreement on small cell vs. large cell morphology: small cell carcinomas were characterized by tumor cells with scant cytoplasm and enlarged, hyperchromatic nuclei with nuclear molding (Supplementary Fig S1A) whereas large cell carcinomas were characterized by tumor cells with more abundant cytoplasm and enlarged nuclei with prominent nucleoli, without nuclear molding (Supplementary Figure S1B). The remaining 15 cases had some degree of overlapping features between small cell and large cell morphology or inter-observer variability (Supplementary Figure S1C).
Most of the cases (11/14 resections; 16/25 overall) had an associated adenoma, adenocarcinoma, or squamous cell carcinoma component. In the right colon, 6/7 (86%) resections or 6/8 (75%) cases overall had either an associated noninvasive adenoma (3/8) or invasive adenocarcinoma (3/8) component. In the left colon, 5/7 (71%) resections or 10/17 (59%) cases overall had an associated noninvasive adenoma/flat dysplasia (6/17), adenocarcinoma (3/17), or squamous cell carcinoma (1/17) component.
p53, Rb, p16, and HPV status of colorectal neuroendocrine carcinomas reveal three molecular patterns
To determine the frequency of TP53 and RB/E2F pathway aberration, we evaluated protein expression of p53, Rb, and p16 by immunohistochemistry and performed PCR and in situ hybridization for HR-HPV (Figs. 1–4, Supplementary Table S3). The 25 neuroendocrine carcinomas could be grouped into one of three major patterns based on Rb expression and HR-HPV status (Fig. 5b).
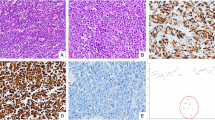
Loss of Rb protein expression in all right colon and a subset of left colon neuroendocrine carcinomas (NEC). This right colon neuroendocrine carcinoma (a−e, case R3) and left colon neuroendocrine carcinoma (f−j, case L4), representatives of the first molecular subgroup in this study, each had an associated tubular adenoma (TA). In both cases, the neuroendocrine carcinoma component showed Rb loss (c, h) and diffuse strong block-like p16 expression (d, i), whereas the TA had intact Rb and negative or wild-type p16. The right colon neuroendocrine carcinoma and associated TA showed wild-type p53 expression (b). The left colon neuroendocrine carcinoma and a subclonal population within the associated TA showed diffuse strong p53 expression (g). Both cases were negative for high-risk human papillomavirus (HR-HPV) by in situ hybridization (ISH). All cases with loss of Rb protein expression also demonstrated genomic alterations in RB1 (see Fig. 4) (Hematoxylin and eosin, a and f)
Intact Rb expression with negative high-risk human papillomavirus (HR-HPV) in a subset of left colon neuroendocrine carcinomas (NEC). This left colon neuroendocrine carcinoma (a−e, L7), representative of the second molecular subgroup in this study, had an associated adenocarcinoma (f−j). Both the neuroendocrine carcinoma and adenocarcinoma components demonstrated diffuse strong p53 (b, g) and intact Rb (c, h). The neuroendocrine carcinoma component had diffuse strong block-like p16 expression (d), whereas the adenocarcinoma had patchy strong p16 expression (i). Both components were negative for HR-HPV by in situ hybridization (ISH) and PCR (e, j). Four left colon neuroendocrine carcinomas with this staining pattern all demonstrated TP53 mutation and 3/4 had focal amplifications of CCNE1, MYC, MYCN, or TERT and 1/4 had SMARCB1 alteration (see Fig. 4) (Hematoxylin and eosin, a and f)
Intact Rb expression with positive high-risk human papillomavirus (HR-HPV) in a subset of left colon neuroendocrine carcinomas (NEC). This left colon neuroendocrine carcinoma (a−e, case L13), representative of the third molecular subgroup in this study, had an associated adenocarcinoma (f−j). Both components demonstrated wild-type p53 expression (b, g), intact Rb (c, h), diffuse strong block-like p16 expression (d, i), and high copy numbers of HR-HPV by in situ hybridization (ISH). None of the cases with this staining pattern had genomic alterations in RB1 or TP53 (see Fig. 4) (Hematoxylin and eosin, a and f)
Genomic alterations in colorectal neuroendocrine carcinomas (NEC). Summary of pathogenic genomic alterations identified in right and left colon neuroendocrine carcinomas (24/25 cases). The top panel shows the presence of a glandular/squamous component; the immunohistochemical (IHC) staining pattern and high-risk human papillomavirus (HR-HPV) status in the neuroendocrine carcinoma component; and any history of inflammatory bowel disease. Group 1 cases, including all right colon neuroendocrine carcinomas (8/8 cases) and 35% of left colon neuroendocrine carcinomas (6/17 cases), had genomic RB1 alterations and were HR-HPV negative. Group 2 cases comprised 24% of left colon neuroendocrine carcinomas (4/17 cases), lacked RB1 alterations, were HR-HPV negative, and showed TP53 alterations combined with focal amplifications in CCNE1, MYC, MYCN, or TERT. Group 3 cases comprised 41% of left colon neuroendocrine carcinomas (7/17, 6 sequenced) had HR-HPV infection and lacked RB1 or TP53 alterations
Rb inactivation is a key driver of colorectal neuroendocrine carcinoma (NEC). a Rb inactivation in colorectal neuroendocrine carcinoma occurs via three distinct mechanisms, which feed into different points within the Rb/E2F signaling pathway. b These three molecular groups can be distinguished by immunohistochemistry (IHC) for Rb and in situ hybridization (ISH) for high-risk human papillomavirus (HR-HPV). Dark gray indicates expression is lost; red indicates diffuse strong expression (p53) or a positive result (HR-HPV). c In the right colon, Rb inactivation was always due to RB1 genomic alteration (mechanism 1). In the left colon, three mutually exclusive mechanisms were observed: (1) RB1 genomic alteration; (2) CCNE1/MYC amplification; and (3) HR-HPV infection
Group 1 (14 cases) demonstrated loss of Rb protein expression and lacked HR-HPV (Figs. 1 and 4, Supplementary Table S3). All 8 (100%) right colon neuroendocrine carcinomas and 6/17 (35%) left colon neuroendocrine carcinomas showed this pattern. Among these 14 cases, 13 (93%) demonstrated diffuse strong block-like expression of p16 and showed loss of Rb. One case had negative p16 and was later found to have a deep deletion of the p16-encoding gene CDKN2A. Half of the cases (7/14) demonstrated aberrant p53 expression (3 negative, 4 diffuse strong). Ten cases had an associated adenoma or adenocarcinoma component, 8 of which had tissue available for comparison with the neuroendocrine carcinoma component. In all eight mixed tumors, Rb expression was intact in the glandular component and lost in the associated neuroendocrine component, although one case demonstrated focal/subclonal loss of Rb in the adenoma (Supplementary Figure S3). The glandular component demonstrated wild-type p16 expression in 7/8 (88%) cases and diffuse strong p16 in 1/8 (12.5%), whereas the synchronous neuroendocrine component always had diffuse strong p16 expression (7/8) or loss of p16 (1/8). Aberrant p53 was seen in both glandular and neuroendocrine components in 5/7 (71%) cases and was restricted to the neuroendocrine component in 2/7 (29%) cases.
Group 2 (4/25 cases) demonstrated intact Rb protein expression and lacked HR-HPV (Figs. 2 and 4, Supplementary Table S3). All four cases with this pattern were in the left colon (three rectal, one rectosigmoid), and two of the four patients had a history of ulcerative colitis. All four demonstrated aberrant p53 expression (one negative, three diffuse strong). Two showed diffuse strong block-like p16 expression, one had p16 loss, and one had patchy strong p16 expression. Three cases had an associated adenoma, flat dysplasia, or adenocarcinoma component. Aberrant p53 expression was observed in both the glandular and neuroendocrine components in 1/3 (33%); the glandular component in the remaining two cases showed a wild-type p53 staining pattern. None of the three mixed tumors demonstrated the same level of p16 expression in the glandular and neuroendocrine components.
Group 3 (7/25 cases) demonstrated high copy numbers of HR-HPV and intact Rb protein expression (Figs. 3 and 4, Supplementary Table S3). All seven of these cases were left-sided (five rectal, two anal). Four out of seven were positive for HPV 18, two for HPV 16, and one for a different high-risk HPV subtype. Only 5/7 cases had sufficient tissue for immunohistochemical studies; of these, all five (100%) demonstrated diffuse strong block-like staining for p16 and a wild-type p53 staining pattern. Three cases had an associated adenoma, adenocarcinoma, or squamous cell carcinoma component, all of which demonstrated identical results in both the non-neuroendocrine and neuroendocrine carcinoma components (positive HR-HPV, block-positive p16, and intact Rb and p53).
All nine rectal neuroendocrine tumors demonstrated wild-type p53 and intact Rb. p16 expression was wild-type in 6/9 (67%), negative in 2/9 (22%), and patchy strong in 1/9 (11%). No cases were associated with HR-HPV (Supplementary Figure S4, Supplementary Table S2). Similarly, none of the 80 cases of colorectal adenocarcinomas were associated with HR-HPV.
Genomic profiling of colorectal neuroendocrine carcinomas reveals three molecular subgroups
To profile the genomic alterations within colorectal neuroendocrine carcinomas, we performed capture-based next-generation sequencing targeting 479 cancer-related genes on 24/25 cases (8 right-sided, 16 left-sided). One case in the left colon that was positive for HR-HPV had insufficient tissue available for sequencing. The sequencing data revealed three molecular subgroups that directly corresponded to those observed by immunohistochemistry (summarized in Fig. 4). All pathogenic and likely pathogenic mutations, focal amplifications, deep deletions, and rearrangements are shown in Fig. 4. Variant details including genomic coordinates and mutant allele frequencies are provided in Supplementary Table S4. Alterations of unknown significance are listed in Supplementary Table S5.
Group 1 cases, which lacked Rb protein and were HR-HPV negative, all demonstrated genomic alterations in RB1. The majority (11/14, 79%) had at least one truncating mutation in RB1 (frameshift, non-sense, or splicing variant) accompanied by either loss of heterozygosity or additional mutations in RB1. The remaining three cases (21%) demonstrated focal deep deletions in RB1. This group also had frequent alterations in TP53 (8/14, 57%), APC (10/14, 71%), and KRAS (5/14, 36%) or BRAF (1/14, 7%). Four of the cases in this group (29%, 3 right colon, 1 left colon) were hypermutated. Two cases in the right colon demonstrated microsatellite instability and were identified as Lynch syndrome patients. The other two hypermutated cases demonstrated POLE p.P286R mutations. These hypermutated cases had numerous truncating variants in additional tumor suppressor genes including KMT2D, PTEN, and NF1 (Fig. 4). Focal amplifications were not common in this group, but one case demonstrated TERT amplification, and another case demonstrated MYCN amplification.
Group 2 cases, which had intact Rb and were HR-HPV negative, had no genomic alterations in RB1, but genomic alteration in other Rb/E2F pathway genes were identified in 2/4 cases. These two cases demonstrated high-level focal amplification in CCNE1 accompanied by high-level focal amplification of MYC or MYCN. The remaining two cases did not show alterations in Rb/E2F pathway genes; one case harbored a truncating variant in SMARCB1 with loss of heterozygosity, and the other case had focal amplification of TERT. All four cases in this group had biallelic alterations in TP53, and two had APC mutations. All four cases were left-sided, and two of the patients had a history of ulcerative colitis.
Group 3 cases, which had intact Rb and were HR-HPV positive, had no genomic alterations in either RB1 or TP53 (6/6 cases). Four (67%) of the cases in this group had alterations in receptor tyrosine kinase, PI3-kinase, or RAS/MAPkinase pathway genes, including ERBB2, PIK3CA, PTEN, and KRAS. The two cases with HPV 16 demonstrated focal high-level SOX2 amplification.
There was no statistically significant difference in overall survival between the three molecular subgroups, which may be due to small sample size. We also compared overall survival between hypermutated and nonhypermutated cases, between tumors arising in the right colon vs. left colon, and between left-sided HPV-related and non-HPV-related tumors. Although none of these reached statistical significance, there was a trend towards better survival outcomes in patients with tumors in the right colon compared with those that had left-sided tumors (hazard ratio 2.56; 95% confidence interval 0.72–9.11; p value = 0.14).
Discussion
The Rb protein is a critical negative regulator of the cell cycle that binds the transcription factor E2F and thus blocks entry into S-phase [34]. In this study, we demonstrate that Rb/E2F pathway dysregulation is near-universal in colorectal neuroendocrine carcinoma and occurs by three distinct molecular mechanisms (Fig. 5). The first two mechanisms involve genomic alteration of either the RB1 gene or other Rb/E2F pathway genes, and both these mechanisms were exclusive to the neuroendocrine carcinoma component of mixed tumors. In 58% of cases, pathway disruption occurred via biallelic alteration of the RB1 gene, either by mutation or deep deletion. These cases showed loss of Rb protein, almost always accompanied by increased p16 expression, except for one case that had a CDKN2A deep deletion. Loss of Rb protein was restricted to the neuroendocrine carcinoma component of all mixed tumors, although we did identify a single case that demonstrated subclonal loss of Rb in a microscopic focus of the associated tubular adenoma (Supplementary Figures S4). These data strongly suggest that inactivation of Rb drives progression of colorectal adenomas or adenocarcinomas to neuroendocrine carcinoma.
A second subset of cases (4/25, all left colon) lacked genomic alteration of RB1 but had alterations in other Rb/E2F pathway genes (2/4 cases). Two cases occurring in ulcerative colitis patients had focal amplifications of CCNE1 and MYC or MYCN, which were accompanied by high p16 expression, again restricted to the neuroendocrine carcinoma component. High-level amplifications are not common in colorectal adenocarcinoma but have been reported to be enriched in patients with inflammatory bowel disease [35, 36]. Another case in this group demonstrated an inactivating mutation in SMARCB1, a core member of the SWI/SNF chromatin remodeling complex. Although SMARCB1 is not considered to be part of the Rb pathway, inactivation of SMARCB1 has been shown to lead to hyperphosphorylation of Rb and upregulation of E2F targets [37]. The last case in this group had a TERT amplification, but no alteration related to Rb was identified.
Finally, the third mechanism of Rb inactivation was mediated by HR-HPV, and this group demonstrated HR-HPV infection in both components of mixed tumors. HPV 18 was the most common viral type detected (4/7) followed by HPV 16 (2/7) and other high-risk strains (1/7). HPV was not detected in any of the 80 colorectal adenocarcinomas used as a control group in our study, which is consistent with prior studies [38]. The HPV oncoprotein E7 interacts with the active, hypophosphorylated form of Rb, which results in both Rb functional inactivation and accelerated degradation [39]. The HPV E6 oncoprotein induces p53 protein degradation [40]. Of note, cases in this group did not show complete loss of Rb or p53 expression by immunohistochemistry, suggesting that these proteins are not fully degraded.
Whereas Rb/E2F pathway dysregulation was seen in 92% (23/25) of cases, p53 inactivation occurred in 76% (19/25) of cases. Forty-eight percent (12/25) had TP53 genomic alterations and 28% (7/25) were HR-HPV positive. The six cases that lacked TP53 genomic alterations or HR-HPV all occurred in molecular group 1 (genomic alteration of RB1), and 50% (3/6) were hypermutated, with dozens of mutations in other tumor suppressor genes (Supplementary Table S4). The fact that Rb but not p53 inactivation is still required in hypermutated cases suggests that Rb may play a more important role in driving neuroendocrine carcinoma than p53.
Interestingly, neuroendocrine carcinoma arising in the right colon occurred exclusively by RB1 genomic alteration, whereas all three mechanisms of Rb inactivation were observed in the left colon. This finding may partly explain the higher frequency of neuroendocrine carcinoma in the latter. These data further support the concept of “sidedness” in colorectal cancer. Recent findings suggest that the optimal first-line treatment for colorectal adenocarcinoma depends on whether the tumor was originally right-sided or left-sided [41, 42]. Our data suggest that the molecular underpinnings of colorectal neuroendocrine carcinoma also depend on “sidedness,” a finding that could have therapeutic implications. Importantly, classification into these three molecular groups ultimately did not require sequencing and could be accomplished in tissue sections using immunohistochemistry and in situ hybridization (Fig. 5b). This approach could potentially alleviate significant challenges in classifying these tumors based on small biopsy material. Although we did not see any statistically significant differences in overall survival between the different molecular groups, we did see a trend towards better outcomes in patients with right-sided tumors compared to those with left-sided tumors. Larger scale genomic and immunohistochemical studies are now needed to determine whether these subgroups are associated with meaningful differences in patient outcomes.
Our findings may have important therapeutic implications. Optimal therapy for colorectal neuroendocrine carcinoma remains ill-defined. Traditionally often treated according to small cell lung cancer guidelines [43], some authors have advocated consideration of adenocarcinoma treatment regimens, in part because colorectal neuroendocrine carcinomas share a subset of molecular alterations with colorectal adenocarcinomas [19]. While our data do show frequent TP53, Wnt pathway, and Ras/MAP-kinase pathway alterations, like colorectal adenocarcinoma, our data highlight Rb/E2F pathway dysregulation as a critical and distinct molecular feature of colorectal neuroendocrine carcinoma that is rarely seen in colorectal adenocarcinoma. This finding supports a requirement for Rb inactivation in colorectal neuroendocrine carcinoma analogous to what has been observed in neuroendocrine carcinoma of the lung and other organs. As such, it remains unclear if the optimal approach to the treatment of colorectal neuroendocrine carcinoma should be organ site-specific or matched to that used for other neuroendocrine carcinomas characterized by Rb loss of function (independent of site of origin) [44,45,46].
Regardless of anatomic site, neuroendocrine carcinoma carries a universally poor prognosis, and platinum-based chemotherapy has served as the mainstay of treatment for both localized and metastatic disease [47]. While most patients initially respond, tumors almost inevitably relapse. Importantly, our sequencing analysis revealed distinct molecular subgroups that may offer new avenues for treatment. Four cases (16%) with RB1 genomic loss also showed a hypermutated phenotype due to either mismatch repair (MMR) deficiency (two cases) or mutation in DNA polymerase epsilon (POLE) (two cases). Patients with MMR-deficient tumors have demonstrated increased clinical benefit from immune checkpoint blockade with the anti-PD-1 antibody pembrolizumab in a phase 2 clinical trial, leading to FDA approval for this indication [48]. This therapeutic response is hypothesized to result from increased tumor neoantigen production that enhances the antitumor immune response [49]. POLE-mutant cancers have an even higher mutation burden, often exceeding 100 mutations per Mb, vs. 10–100 per Mb in MMR-deficient tumors and 1–10 per Mb in microsatellite stable tumors [50]. Mutations in the proofreading exonuclease domains of POLE and DNA polymerase delta 1 (POLD1) have been identified in patients with non-small cell lung cancer and endometrial carcinoma who responded to pembrolizumab [51, 52]. Nevertheless, most POLE-mutant tumors are not detected by routine microsatellite instability testing. Currently, a phase 2 clinical trial of pembrolizumab at our institution is limited to patients with previously treated extrapulmonary neuroendocrine carcinoma (NCT03136055). Given the potential for significant therapeutic benefit from immunotherapy, further testing for a hypermutated phenotype in colorectal neuroendocrine carcinoma may be indicated to identify candidate patients who would benefit from this treatment approach.
Finally, our study highlights colorectal neuroendocrine carcinoma as another tumor type that is driven by HR-HPV, which has important implications for screening, prevention, and treatment. In terms of treatment, HPV-driven neuroendocrine carcinoma may also respond to immune checkpoint blockade, even without a hypermutated phenotype. For example, response to pembrolizumab has been reported in advanced Merkel-cell carcinoma, both in tumors positive for the Merkel-cell polyomavirus and those due to ultraviolet light, which have markedly different mutational burdens [53]. Despite having a low mutational load, virus-associated tumors are thought to induce an antitumor immune response via expression of viral antigens. Similarly, HPV-associated carcinoma may elicit an HPV-specific immune response, and this rationale has recently motivated multiple clinical trials for immunotherapy in advanced cervical cancer [54].
In summary, we have identified a critical role for Rb in the molecular pathogenesis of colorectal neuroendocrine carcinoma and found that a significant subset of rectal and anal tumors is driven by HR-HPV infection. Future studies are indicated to evaluate the frequency of both virally driven and hypermutated tumors and whether distinct molecular subgroups of neuroendocrine carcinoma may benefit from specifically targeted therapeutic approaches.
References
Perren A, Couvelard A, Scoazec JY, Costa F, Borbath I, Delle Fave G, et al. ENETS Consensus Guidelines for the Standards of Care in Neuroendocrine Tumors: Pathology: Diagnosis and Prognostic Stratification. Neuroendocrinology. 2017;105:196–200.
Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71.
George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53.
Tang LH, Basturk O, Sue JJ, Klimstra DS. A Practical Approach to the Classification of WHO Grade 3 (G3) Well-differentiated Neuroendocrine Tumor (WD-NET) and Poorly Differentiated Neuroendocrine Carcinoma (PD-NEC) of the Pancreas. Am J Surg Pathol. 2016;40:1192–202.
Tang LH, Untch BR, Reidy DL, O'Reilly E, Dhall D, Jih L, et al. Well-Differentiated Neuroendocrine Tumors with a Morphologically Apparent High-Grade Component: A Pathway Distinct from Poorly Differentiated Neuroendocrine Carcinomas. Clin Cancer Res. 2016;22:1011–7.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol. 2017;35:3065–74.
Rickman DS, Beltran H, Demichelis F, Rubin MA. Biology and evolution of poorly differentiated neuroendocrine tumors. Nat Med. 2017;23:1–10.
Dasari A, Mehta K, Byers LA, Sorbye H, Yao JC. Comparative study of lung and extrapulmonary poorly differentiated neuroendocrine carcinomas: A SEER database analysis of 162,983 cases. Cancer. 2018;124:807–15.
Oberg K. The genetics of neuroendocrine tumors. Semin Oncol. 2013;40:37–44.
Kloppel G, Klimstra DS, Hruban RH, Adsay V, Capella C, Couvelard A, et al. Pancreatic Neuroendocrine Tumors: Update on the New World Health Organization Classification. Ajsp-Reviews and Reports. 2017;22:233–9.
Stoler MH, Mills SE, Gersell DJ, Walker AN. Small-cell neuroendocrine carcinoma of the cervix. A human papillomavirus type 18-associated cancer. Am J Surg Pathol. 1991;15:28–32.
Bishop JA, Westra WH. Human papillomavirus-related small cell carcinoma of the oropharynx. Am J Surg Pathol. 2011;35:1679–84.
Kraft S, Faquin WC, Krane JF. HPV-associated neuroendocrine carcinoma of the oropharynx: a rare new entity with potentially aggressive clinical behavior. Am J Surg Pathol. 2012;36:321–30.
Cimino-Mathews A, Sharma R, Illei PB. Detection of human papillomavirus in small cell carcinomas of the anus and rectum. Am J Surg Pathol. 2012;36:1087–92.
zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50.
Bernick PE, Klimstra DS, Shia J, Minsky B, Saltz L, Shi W, et al. Neuroendocrine carcinomas of the colon and rectum. Dis Colon Rectum. 2004;47:163–9.
Takizawa N, Ohishi Y, Hirahashi M, Takahashi S, Nakamura K, Tanaka M, et al. Molecular characteristics of colorectal neuroendocrine carcinoma; similarities with adenocarcinoma rather than neuroendocrine tumor. Hum Pathol. 2015;46:1890–900.
Jesinghaus M, Konukiewitz B, Keller G, Kloor M, Steiger K, Reiche M, et al. Colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas are genetically closely related to colorectal adenocarcinomas. Mod Pathol. 2017;30:610–9.
Woischke C, Schaaf CW, Yang HM, Vieth M, Veits L, Geddert H, et al. In-depth mutational analyses of colorectal neuroendocrine carcinomas with adenoma or adenocarcinoma components. Mod Pathol. 2017;30:95–103.
Darragh TM, Colgan TJ, Cox JT, Heller DS, Henry MR, Luff RD, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-Associated Lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205–42.
Kerr DA, Sweeney B, Arpin RN, Ring M, Pitman MB, Wilbur DC, et al. Automated Extraction of Formalin-Fixed, Paraffin-Embedded Tissue for High-Risk Human Papillomavirus Testing of Head and Neck Squamous Cell Carcinomas Using the Roche Cobas 4800 System. Arch Pathol Lab Med. 2016;140:844–8.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–9.
Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:1–33.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71.
Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM: Broad Institute. http://broadinstitute.github.io/picard.
Garrison E, Marth, G Haplotype-based variant detection from short-read sequencing. arXiv 2012;1207.3907 [q-bio.GN].
Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–7.
Yaeger R, Shah MA, Miller VA, Kelsen JR, Wang K, Heins ZJ, et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology. 2016;151:278–87, e6.
Fujita M, Matsubara N, Matsuda I, Maejima K, Oosawa A, Yamano T, et al. Genomic landscape of colitis-associated cancer indicates the impact of chronic inflammation and its stratification by mutations in the Wnt signaling. Oncotarget. 2018;9:969–81.
Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci USA. 2005;102:17745–50.
Gornick MC, Castellsague X, Sanchez G, Giordano TJ, Vinco M, Greenson JK, et al. Human papillomavirus is not associated with colorectal cancer in a large international study. Cancer Causes Control. 2010;21:737–43.
Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4.
Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36.
Tejpar S, Stintzing S, Ciardiello F. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials (vol 3, pg 194, 2016). JamaOncol. 2017;3:194–201.
Venook AP, Niedzwiecki D, Innocenti F, Fruth B, Greene C, O'Neil BH, et al. Impact of primary (1 degrees) tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol. 2016;34:3504.
Network NCC. NCCN Clinical Practice Guidelines in Oncology for Neuroendocrine and Adrenal Tumors (Version 2.2018). https://www.nccn.org/professionals/physician_gls/pdf/neuroendocrine.pdf. Accessed: May 6, 2018.
Smith J, Reidy-Lagunes D. The management of extrapulmonary poorly differentiated (high-grade) neuroendocrine carcinomas. Semin Oncol. 2013;40:100–8.
Sorbye H, Strosberg J, Baudin E, Klimstra DS, Yao JC. Gastroenteropancreatic high-grade neuroendocrine carcinoma. Cancer. 2014;120:2814–23.
Sorbye H, Welin S, Langer SW, Vestermark LW, Holt N, Osterlund P, et al. Predictive and prognostic factors for treatment and survival in 305 patients with advanced gastrointestinal neuroendocrine carcinoma (WHO G3): the NORDIC NEC study. Ann Oncol. 2013;24:152–60.
Bergsland EK. The evolving landscape of neuroendocrine tumors. Semin Oncol. 2013;40:4–22.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20.
Nebot-Bral L, Brandao D, Verlingue L, Rouleau E, Caron O, Despras E, et al. Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer. 2017;84:290–303.
Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–50.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8.
Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126:2334–40.
Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med. 2016;374:2542–52.
Borcoman E, Le Tourneau C. Pembrolizumab in cervical cancer: latest evidence and clinical usefulness. Ther Adv Med Oncol. 2017;9:431–9.
Acknowledgements
This study was funded by the University of California San Francisco Department of Pathology Research Endowment award to N.M.J. The authors would like to thank the pathology departments of the hospitals that provided slides and blocks for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Shamir, E.R., Devine, W.P., Pekmezci, M. et al. Identification of high-risk human papillomavirus and Rb/E2F pathway genomic alterations in mutually exclusive subsets of colorectal neuroendocrine carcinoma. Mod Pathol 32, 290–305 (2019). https://doi.org/10.1038/s41379-018-0131-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0131-6
This article is cited by
-
Hsa-miR-194-5p and hsa-miR-195-5p are down-regulated expressed in high dysplasia HPV-positive Pap smear samples compared to normal cytology HPV-positive Pap smear samples
BMC Infectious Diseases (2024)
-
Aktuelle WHO-Klassifikation (2022) neuroendokriner Neoplasien
Die Radiologie (2024)
-
Molecular Classification of Gastrointestinal and Pancreatic Neuroendocrine Neoplasms: Are We Ready for That?
Endocrine Pathology (2024)
-
Neuroendocrine neoplasms of the lung and gastrointestinal system: convergent biology and a path to better therapies
Nature Reviews Clinical Oncology (2023)
-
Potent molecular-targeted therapies for gastro-entero-pancreatic neuroendocrine carcinoma
Cancer and Metastasis Reviews (2023)