Abstract
Targeted inhibitors of neurotropic tyrosine kinases are highly effective in selected patients with gene fusions involving NTRK1, NTRK2, or NTRK3. These fusions are consistently detected in rare cancer types (e.g., secretory breast carcinoma and congenital infantile fibrosarcoma), but the occurrence of NTRK fusions in common cancers and their relationship to other therapy biomarkers are largely unexplored. Tissue samples from 11,502 patients were analyzed for 53 gene fusions and sequencing of 592 genes, along with an immunohistochemical evaluation of TrkA/B/C and PD-L1. Thirty-one cases (0.27% of the entire cohort) had NTRK fusions. The most common fusions were ETV6:NTRK3 (n = 10) and TPM3:NTRK1 (n = 6). Gliomas had the highest number of NTRK fusions (14/982, 1.4%), most commonly involving NTRK2 (n = 9). Seventeen non-glioma cases with NTRK fusions included carcinomas of the lungs, thyroid, breast, cervix, colon, nasal cavity, cancer of unknown primary and soft tissue sarcomas. Strong and uniform Trk expression detected with a pan-Trk immunohistochemistry characterized 7/8 NTRK1 fusion cases and 8/9 NTRK2 fusion cases, while NTRK3 fused cases were positive in 6/11 (55%) of cases. 29% of NTRK fusion cases had no other pathogenic genomic alteration. PD-L1 expression was observed in 23% of NTRK fused cases while high tumor DNA microsatellite instability was detected in two cases. We confirm the rarity of NTRK genes fusions outside the brain malignancies. NTRK inhibitors alone or combined with immune checkpoint inhibitors may be a therapeutic option for a substantial proportion of these patients. Strategies for detection of the NTRK fusion-driven cancers may include immunohistochemistry, but gene fusion detection remains the most reliable tool.
Similar content being viewed by others
Introduction
The tropomyosin receptor kinase (Trk) family is composed of three transmembrane protein receptors TrkA, TrkB, and TrkC that are encoded by the NTRK1, NTRK2, and NTRK3 genes, respectively [1, 2]. They are actively involved in neuronal development, maintenance, and protection [1,2,3]. Gene fusions involving NTRK1, 2, and 3 and their partner genes result in a constitutive activation or overexpression of Trk receptors, potentially leading to oncogenesis [2]. These fusion proteins may induce cancer cell proliferation and activate downstream signaling pathways (PI3K, RAS/MAPK/ERK, PLC-gamma).
Due to their potential oncogenic activity, NTRK fusions have been actively investigated as therapeutic targets [1, 2]. Therapies targeting NTRKs include larotrectinib (or LOXO-101; selective inhibitor of Trk) and entrectinib (pan-Trk, ROS1, and ALK inhibitor) and have shown a promising activity in various cancer types harboring NTRK fusions [1, 2, 4,5,6,7,8,9,10,11,12,13,14,15,16].
NTRK fusions are consistently detected in rare types of cancers (secretory breast carcinoma, mammary analogue secretory carcinoma, congenital infantile fibrosarcoma, and congenital mesoblastic nephroma) as well as in various pediatric cancers (Supplemental Table 1) [4, 17,18,19]. A small percentage of common adult patients’ cancers (non-small cell lung cancer, salivary gland, colorectal, head and neck, thyroid, bladder cancers as well as malignant melanomas, soft tissue sarcomas and brain tumors [gliomas]) carry fusions of NTRK genes [1, 2, 4, 8, 20,21,22,23,24,25,26].
NTRK gene fusions are usually detected by various molecular assays such as next-generation sequencing of DNA, or targeted RNA and fluorescence in-situ hybridization [21]. Limited data are available regarding the diagnostic utility of immunohistochemistry in detecting NTRK gene alterations in oncologic pathology practice [21, 22, 27, 28].
In the present study, we reviewed a large cohort of solid malignancies profiled by a commercial laboratory (Caris Life Sciences, Phoenix, AZ) that were tested for NTRK gene fusions and other pathogenic/targetable genomic and protein alterations. Additionally, we assessed a diagnostic utility of immunohistochemistry in detecting NTRK gene fusions.
Materials and methods
Samples
The study included 11,502 formalin-fixed paraffin-embedded tissue samples of various solid cancer types that have been profiled at Caris Life Sciences in the period 2015–2018. The majority of the samples (>90%) submitted for molecular profiling at Caris originated from pathology laboratories in the US. Prior to molecular testing, all cases were reviewed by a board-certified pathologist to confirm the diagnosis and select appropriate slides/tissue areas for testing.
Next-generation sequencing and Archer Fusion assay
The cases were analyzed with massively parallel next-generation sequencing platform (592-Gene Panel utilizes SureSelect XT biotinylated RNA probes from Agilent, Santa Clara, CA, to capture DNA fragments from the exons of 592 genes; sequencing was performed using NextSeq instrument from Illumina, San Diego, CA). The total mutational burden was calculated by counting nonsynonymous missense mutations while excluding common germline variants. Tumor mutational burden was considered high when ≥17 mutations/megabase were present [29]. Microsatellite instability was calculated from the next-generation sequencing data by direct analysis of short tandem repeat tracts in the target regions of sequenced genes. The count only included alterations that resulted in increases or decreases in the number of repeats; high microsatellite instability was defined as ≥46 altered microsatellite loci (this threshold was established by comparing next-generation sequencing to the polymerase chain reaction-based microsatellite fragments analysis results from ~2100 cases) [30].
ArcherDx FusionPlex Assay (ArcherDX, Boulder, CO) was used to detect gene fusions (53 gene targets in total including NTRK1, NTRK2, and NTRK3 genes). The complete panel of tested gene fusions is available at https://www.carismolecularintelligence.com/tumor-profiling-menu/mi-profile-usa-excluding-new-york/.
Immunohistochemistry
Pan-Trk immunohistochemical staining for TrkA/B/C expression was performed using pan-Trk antibody (rabbit recombinant monoclonal antibody, clone EPR17341, Abcam, Cambridge, MA). Tumors were considered positive if ≥1% of tumor cells exhibited positivity at any intensity above background. Different subcellular staining patterns were considered positive, as previously suggested (cytoplasmic, membranous, nuclear, and perinuclear) [21]. PD-L1 expression was evaluated in the tumor cells using either SP142 antibody (Ventana; for non-lung carcinoma cases) or 22c3 clones (Agilent DAKO, for non-small cell lung carcinoma cases). Tumor cells were considered positive for SP142 PD-L1 if ≥5% of cancer cells exhibited moderate (2+) membranous positivity [30,31,32,33], while 22c3 positivity in non-small cell lung carcinomas was determined based on companion diagnostics criteria (https://www.agilent.com/cs/library/usermanuals/public/29158_pd-l1-ihc-22C3-pharmdx-nsclc-interpretation-manual.pdf). Placental tissue served as a positive control for PD-L1 antibodies, while cerebral cortical tissue was used for pan-Trk controls.
Immunohistochemical stains were performed using automated platforms (Benchmark, Ventana Medical Systems, Inc. and DAKO Autostainer, Agilent). All assays were performed in CLIA/CAP/ISO15189/NYSDOH certified clinical laboratory (Caris Life Sciences, Phoenix, AZ).
Results
Patients and samples’ characteristics
The study included samples from adult patients (mean age: 55 years; range: 11–90 years) with the exception of one pediatric patient with glioblastoma multiforme (an 11-year-old male).
The tissue distribution of studied cancers with NTRK fusions included non-small cell lung carcinomas (n = 4073), colorectal carcinomas (n = 1272), gliomas (n = 982), breast carcinomas (n = 769), various soft tissue sarcomas (n = 478), cancers of unknown primary (n = 227), thyroid carcinomas (n = 70), cervical carcinomas (n = 68) (Fig. 2). Cancers of various other primary sites without NTRK fusions (>3000) were included for comparison.
Fusion assay results for NTRKs
The fusion assay identified 168 cases with a validated fusion of any type (1.5% for the entire cohort) of which 31 (0.27% of the entire cohort) had a NTRK gene fusion (Table 1, Fig. 2, Supplemental Table 2). No case had more than one fusion detected. Three NTRK genes were involved in fusions with 15 different gene partners; ETV6:NTRK3 and TPM3:NTRK1 were the most common fusions (10 cases and 6 cases, respectively) (Table 1; Figs. 1 and 2; Supplemental Table 2).
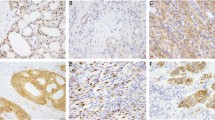
a TrkA protein expression was detected in nuclear, cytoplasmic, and membranous pattern in a case of lung adenocarcinoma. b A weak (1+), sporadic TrkB expression (cytoplasmic pattern) in lung adenocarcinoma with NTRK2 fusion (pleural effusion). c Nuclear expression of TrkC in a case of carcinoma of unknown primary with NTRK3 gene fusion. d A strong (3+ intensity) and diffuse (100%) cytoplasmic expression of TrkA in a case of poorly differentiated colorectal carcinoma with microsatellite instability status caused by the loss of MSH6 protein. Interestingly, this case also exhibited a diffuse and strong PD-L1 expression (not shown)
Four thyroid carcinomas (5.7%) (two poorly differentiated, one papillary, and one anaplastic thyroid carcinoma) harbored the highest proportion of the NTRK fusions while gliomas had the highest number of NTRK fusions involving all three NTRK genes (14/982 cases, 1.4%), most commonly NTRK2 (nine cases fused with eight different genes). Seventeen non-glioma cases with NTRK fusions included lung adenocarcinoma (n = 4), thyroid carcinoma (n = 4), colorectal carcinoma (n = 2), cancers of unknown primary site (n = 2), sarcomas (one soft tissue and one uterine sarcoma, not otherwise specified), cervix, breast secretory carcinoma, and sinonasal adenocarcinoma (non-intestinal type) (n = 1, each).
Immunohistochemistry for Trk proteins
To explore the diagnostic utility of pNTRK immunohistochemistry, we analyzed 4136 cases including 28 confirmed NTRK fusion-positive cancers. The results of the concordance between Archer Fusion and immunohistochemical assays are presented in Table 2. Strong (3+) and uniform (100% of cancer cells) staining characterized 7/8 NTRK1 fused cases (5/6 TPM3, 1/1 BCAN, and 1/1 MEF2D case) (Fig. 1a). The subcellular distribution of the immunohistochemical stain was cytoplasmic and/or membranous (Fig. 1a, d). NTRK2 gene fusion cases were variably positive in 8/9 cases with cytoplasmic and perinuclear distribution (Fig. 1b), while NTRK3 gene fusion cases (nuclear and cytoplasmic/membranous; Fig. 1c) were negative in 5/11 cases including 4 out of 9 ETV6–NTRK3 positive cases. Taken together, these results yielded overall sensitivity 75% and specificity 95.9%. Positive predictive value and negative predictive value were 11.2% and 99.8%, respectively.
Other co-occurring molecular genomic alterations in NTRK fused cases
Of 31 NTRK fusion-positive cases, nine (29%) had no other pathogenic alteration (gene mutations or amplifications) (Supplemental Table 2). The tumor mutational burden varied between 1 and 37 somatic mutations per megabase and was the highest in two microsatellite instability high cases (colorectal carcinoma case: TPM3–NTRK1 and cancer of unknown primary case: ETV6–NTRK3). Additionally, PD-L1 expression was detected in 7/30 cases (23%) including both microsatellite instability high cases (Supplemental Table 1). The most frequent mutations seen in concurrence with NTRK fusions were TP53, PTEN, and PIK3CA mutations. Targetable alterations included EGFR and MET amplification (1 case) and VEGFR2 gene amplification and mutation (1 case) (Supplemental Table 2).
Discussion
In the present study, we screened a large number of common solid cancers in adult patients for the presence of NTRK gene fusions confirming the rarity of such events across various histologic cancer types. The data obtained in our study are generally in line with previous studies [1, 2, 4, 20,21,22,23, 34, 35], but offer a much broader overview on the frequency and types of NTRK fusions given the large sample size (>11,000 patients screened). We detected NTRK fusions in a wide variety of histologic tumor types, predominantly carcinomas and brain gliomas. We also report the detection of NTRK fusions in two undifferentiated sarcomas: one in soft tissues (TPM3:NTRK1) and the other arising in the uterus (SPECC1L:NTRK3). This is in line with a recent report of Chiang et al. who described a uterine spindle cell sarcoma with an NTRK fusion, resembling fibrosarcoma and distinct from uterine leiomyosarcomas [24].
The identification of NTRK fusions has recently come into focus due to the availability of targeted treatment modalities (NTRK inhibitors). Initial data exploring NTRK-rearranged cancers treated with the two most commonly used NTRK inhibitors entrectinib and/or larotrectinib demonstrated a remarkable antitumor activity with acceptable toxicity profile; some patients achieved rapid and durable therapeutic responses [2, 4, 5, 9, 14,15,16, 36, 37]. Drilon et al. demonstrated that 86% of the patients treated with larotrectinib were continuing treatment with the NTRK inhibitor or had undergone curative surgical treatment (a median follow-up was 9.4 months) [4]. In addition, there are ongoing Phase 2 trials with entrectinib and larotrectinib involving the patients whose cancers harbor NTRK gene fusions, irrespective of tumor histology (“basket trials”) as well as Phase 1 trials with other NTRK inhibitors (MGCD516, PLX7486, DS-6051b, TSR-011) [2, 16]. One of the important additional findings in our study is the fact that 29% of detected NTRK fused cancers had no other pathogenic genomic alterations detected by the 592-gene next-generation sequencing panel. This makes the detection of NTRK fusions even more therapeutically relevant [35].
In order to maximize the detection of patients with tumors carrying targetable alterations, numerous studies have demonstrated that immunohistochemistry may serve as an effective screening tool for various cancer types before highly sensitive, but less available molecular tests such as fluorescent in-situ hybridization or next-generation sequencing were performed. Thus, ALK (D5F3) immunohistochemistry-based test for ALK gene alterations (fusions) has been approved by the Food and Drug Administration as a companion diagnostics for the treatment of non-small cell lung cancer patients with crizotinib and other ALK inhibitors [38, 39]. Reports also support the potential diagnostic utility of ROS1 immunohistochemistry for detection of ROS1 gene rearrangements in non-small cell lung cancer patients [40, 41]. Limited data are available regarding the screening of NTRK fusions by immunohistochemistry [21, 27, 28]. Our results (Table 2) indicate that an immunohistochemistry assay using a pan-Trk antibody may help in the detection of NTRK1 and NTRK2 gene fusions, but may not be sensitive enough for the detection of NTRK3 gene fusions (~45% of these cases had false negative results). Our study found immunohistochemistry to be overall less sensitive (75%) than the study of Hechtman et al. (sensitivity 95.2% and specificity 100%) [21] and the study of Rudzinski et al. [Pan-Trk IHC (EPR17341) was 97% sensitive and 98% specific for NTRK rearrangements] [42]. A study of Murphy et al. [27] revealed that the pan-Trk immunohistochemical test had 100% negative predictive value for NTRK gene fusions; however, among the immunohistochemically positive cases, only 9% harbored NTRK gene fusions (prevalence was 45%). A recent study of Hung et al. [28] confirmed a high sensitivity but not high specificity of pan-Trk immunohistochemistry in identifying infantile fibrosarcomas with NTRK3 gene fusions.
In conclusion, NTRK gene fusions are rare in common solid cancers but may be present with an increased frequency in a proportion of some highly aggressive cancers (e.g., glioblastoma multiforme). NTRK fusions may be detected as the only actionable target or co-occur with other pathogenic and/or potentially targetable alterations (e.g., immune checkpoint inhibitors for PD-L1 positive or microsatellite instability-high cases), which opens the opportunity for combination therapies for future investigations. Concurrent alterations observed in single cases with MET and VEGFR2 encourage the use of multi-targeted therapies including cabozantinib, merestinib, or MGCD516, warrants further studies. Immunohistochemistry for Trk proteins may serve as a screening tool for detection of NTRK gene fusions, particularly NTRK1 and NTRK2 fusions, but improved antibodies are necessary to detect all cases with NTRK gene fusions.
References
Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1:e000023.
Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58–66.
Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107–14.
Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–9.
Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 STARTRK-1). Cancer Discov. 2017;7:400–9.
Nagasubramanian R, Wei J, Gordon P, Rastatter JC, Cox MC, Pappo A. Infantile fibrosarcoma with NTRK3–ETV6 fusion successfully treated with the tropomyosin-related kinase inhibitor LOXO-101. Pediatr Blood Cancer. 2016;63:1468–70.
Drilon A, Li G, Dogan S, Gounder M, Shen R, Arcila M, et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6–NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol. 2016;27:920–6.
Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5:1049–57.
Rolfo C, Raez L. New targets bring hope in squamous cell lung cancer: neurotrophic tyrosine kinase gene fusions. Lab Invest. 2017;97:1268–70.
Rolfo C, Ruiz R, Giovannetti E, Gil-Bazo I, Russo A, Passiglia F, et al. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin Investig Drugs. 2015;24:1493–500.
Sigal D, Tartar M, Xavier M, Bao F, Foley P, Luo D, et al. Activity of entrectinib in a patient with the first reported NTRK fusion in neuroendocrine cancer. J Natl Compr Cancer Netw. 2017;15:1317–22.
Smith KM, Fagan PC, Pomari E, Germano G, Frasson C, Walsh C, et al. Antitumor activity of entrectinib, a pan-TRK, ROS1, and ALK inhibitor, in ETV6–NTRK3-positive acute myeloid leukemia. Mol Cancer Ther. 2018;17:455–63.
Farago AF, Le LP, Zheng Z, Muzikansky A, Drilon A, Patel M, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer.J Thorac Oncol. 2015;10:1670–4.
Sidaway P. Targeted therapy: larotrectinib effective against TRK-fusion-positive cancers. Nat Rev Clin Oncol. 2018;10:1670–4.
Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19:705–14.
Lange AM, Lo HW. Inhibiting TRK proteins in clinical cancer therapy. Cancers (Basel). 2018;10:E105.
Davis JL, Lockwood CM, Albert CM, Tsuchiya K, Hawkins DS, Rudzinski ER. Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol. 2018;21:68–78.
Pavlick D, Schrock AB, Malicki D, Stephens PJ, Kuo DJ, Ahn H, et al. Identification of NTRK fusions in pediatric mesenchymal tumors. Pediatr Blood Cancer. 2017;64.
Prasad ML, Vyas M, Horne MJ, Virk RK, Morotti R, Liu Z, et al. NTRK fusion oncogenes in pediatric papillary thyroid carcinoma in northeast United States. Cancer. 2016;122:1097–107.
Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–54.
Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41:1547–51.
Lee SJ, Li GG, Kim ST, Hong ME, Jang J, Yoon N, et al. NTRK1 rearrangement in colorectal cancer patients: evidence for actionable target using patient-derived tumor cell line. Oncotarget. 2015;6:39028–35.
Church AJ, Calicchio ML, Nardi V, Skalova A, Pinto A, Dillon DA, et al. Recurrent EML4–NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol. 2018;31:463–73.
Chiang S, Cotzia P, Hyman DM, Drilon A, Tap WD, Zhang L, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42:791–8.
Landman Y, Ilouze M, Wein S, Neiman V, Yerushalmi R, Yakimov M, et al. Rapid response to larotrectinib (LOXO-101) in an adult chemotherapy-naive patients with advanced triple-negative secretory breast cancer expressing ETV6–NTRK3 fusion. Clin Breast Cancer. 2017;18:e267–70.
Lezcano C, Shoushtari AN, Ariyan C, Hollmann TJ, Busam KJ. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol. 2018;42:1052–8.
Murphy DA, Ely HA, Shoemaker R, Boomer A, Culver BP, Hoskins I, et al. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol. 2017;25:513–23.
Hung YP, Fletcher CDM, Hornick JL. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumor, and histologic mimics. Histopathology. 2018.
Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746–56.
Gatalica Z, Xiu J, Swensen J, Vranic S. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur J Cancer. 2018;94:179–86.
Carbognin L, Pilotto S, Milella M, Vaccaro V, Brunelli M, Calio A, et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS One. 2015;10:e0130142.
Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–62.
Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–74.
Bastos AU, de Jesus AC, Cerutti JM. ETV6–NTRK3 and STRN-ALK kinase fusions are recurrent events in papillary thyroid cancer of adult population. Eur J Endocrinol. 2018;178:85–93.
Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–72.
Ardini E, Menichincheri M, Banfi P, Bosotti R, De Ponti C, Pulci R, et al. Entrectinib, a pan-TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol Cancer Ther. 2016;15:628–39.
Menichincheri M, Ardini E, Magnaghi P, Avanzi N, Banfi P, Bossi R, et al. Discovery of entrectinib: a new 3-aminoindazole as a potent anaplastic lymphoma kinase (ALK), c-ros oncogene 1 kinase (ROS1), and pan-tropomyosin receptor kinases (pan-TRKs) inhibitor. J Med Chem. 2016;59:3392–408.
Thorne-Nuzzo T, Williams C, Catallini A, Clements J, Singh S, Amberson J, et al. A sensitive ALK immunohistochemistry companion diagnostic test identifies patients eligible for treatment with crizotinib. J Thorac Oncol. 2017;12:804–13.
Blackhall FH, Peters S, Bubendorf L, Dafni U, Kerr KM, Hager H, et al. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape Project. J Clin Oncol. 2014;32:2780–7.
Sholl LM, Sun H, Butaney M, Zhang C, Lee C, Janne PA, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol. 2013;37:1441–9.
Cha YJ, Lee JS, Kim HR, Lim SM, Cho BC, Lee CY, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One. 2014;9:e103333.
Rudzinski ER, Lockwood CM, Stohr BA, Vargas SO, Sheridan R, Black JO, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol. 2018;42:927–35.
Acknowledgements
The preliminary data from this study were presented at the AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics: Discovery, Biology, and Clinical Applications; October 26–30, 2017, Philadelphia, PA, USA. We are thankful to Dr. Todd Maney for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Zoran Gatalica, Joanne Xiu, and Jeffrey Swensen are all employees of the Caris Life Sciences. Semir Vranic has received honoraria from Caris Life Sciences.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Gatalica, Z., Xiu, J., Swensen, J. et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 32, 147–153 (2019). https://doi.org/10.1038/s41379-018-0118-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0118-3
This article is cited by
-
Validation and interpretation of Pan-TRK immunohistochemistry: a practical approach and challenges with interpretation
Diagnostic Pathology (2024)
-
NTRK-rearranged spindle cell sarcoma of the uterine cervix with a novel NUMA1::NTRK1 fusion
Virchows Archiv (2024)
-
Evaluation of NTRK expression and fusions in a large cohort of early-stage lung cancer
Clinical and Experimental Medicine (2024)
-
Characterization of the Rhizosphere Bacterial Microbiome and Coffee Bean Fermentation in the Castillo-Tambo and Bourbon Varieties in the Popayán-Colombia Plateau
BMC Plant Biology (2023)
-
Diffuse pulmonary meningotheliomatosis with pan-TRK expression by immunohistochemistry: a novel finding and potential pitfall
Diagnostic Pathology (2023)