Abstract
In this study, we evaluate the expression of human papillomavirus E4 protein (marker for the onset of a productive infection) and hypermethylation of host-cell CADM1, MAL, and miR124-2 genes (marker for an advanced, transforming infection) in cervical intraepithelial neoplasia (CIN) and cancer. A total of 115 cervical lesions were categorized by 3 pathologists into no dysplasia, CIN1, CIN2, CIN3, or cancer by classical histomorphological grading criteria, and by an immunoscore (cumulative value: 0–6) grading system based on Ki-67 (score: 0–3) and p16ink4a (score: 0–3) expression. Lesions were immunostained for E4 protein and analyzed for hypermethylation of CADM1, MAL, or miR124-2 genes. Expression of E4 and hypermethylation levels were related to CIN grade based on both classical and immunoscore grading. Hypermethylation increased with severity of the lesion as defined by both classical histomorphological grading and immunoscore criteria, and was always present in carcinomas (22/22). Extensive E4 expression decreased with increasing CIN grade and immunoscore, being most frequent in classically graded CIN1 or in lesions with cumulative immunoscore 1–3 and absent in carcinomas. High-grade lesions (CIN2/3 or immunoscore: 4–6) showed less E4 expression, which was inversely related to an increasing hypermethylation. Extensive E4 expression, as observed in a small proportion of high-grade lesions (6/49 and 8/43, respectively), was mostly associated with a negative methylation marker status (5/6 and 7/8, respectively). Our results illustrate the gradual transition of productive CIN (reflected by extensive E4 expression), to advanced transforming CIN (reflected by extensive hypermethylation) and cancer. Expression patterns of E4 and hypermethylation status of host-cell genes, may be used to identify cervical lesions at risk for cervical cancer, providing a better guidance for clinicians on treatment decisions.
Similar content being viewed by others
Introduction
Cervical cancer screening programs aim to detect cervical cancer at an early or precancerous stage, so-called cervical intraepithelial neoplasia (CIN). The grade of CIN is currently defined by the extent of the dysplastic cells and the severity of cellular abnormalities in the squamous epithelium, and is based on morphological features in hematoxylin and eosin (H&E) stained sections either with or without aid of adjunct immunohistochemical (IHC) stainings [1]. CIN3 lesions are generally considered direct precursors of cervical cancer and in need of treatment, whereas for CIN1 lesions strict follow-up is considered sufficient as a clinical management strategy. The management of CIN2 is diverse. CIN2 constitute a heterogeneous group, which can either be the result of a productive or a transforming human papillomavirus (HPV) infection, which have distinct cancer risks [2,3,4,5]. Moreover, there is moderate reproducibility of CIN grading, especially for CIN2 [6–9]. These diversities result in the current suboptimal situation, wherein cervical lesions with a similar cancer progression risk are diagnosed and managed differently, while cervical lesions with a distinct cancer progression risk are diagnosed and managed similarly. This leads to overtreatment of CIN lesions with a low short-term progression risk, which is specifically critical among young women due to the influence of excision of the transformation zone on fertility rates and pregnancy outcomes [10]. The number of women unnecessarily treated can be reduced by even small improvements in accuracy of the diagnosis of CIN, identifying CIN with a high short-term cancer progression risk [4].
Grading of CIN can be optimized by the use of Ki-67 and p16ink4a immunostainings. In terms of accuracy and reproducibility, we recently showed that a cumulative score value (immunoscore) based solely on a three-tiered scoring system for both Ki-67 (score: 0–3) and p16ink4a (score: 0–3) seemed most optimal [6]. In particular, the detection of CIN3 (treatment) and CIN1 (no treatment) has been shown to be more accurate and less variable by use of this grading system. However, to further improve the identification of cervical precursor lesions in need for treatment, additional biomarkers reflecting the cancer risk are necessary. The HPV E4 protein is a marker for the onset of a productive HPV infection and particularly has been shown to be present in a high proportion of CIN2, but not in most CIN3 [11,12,13]. Promoter hypermethylation of host-cell genes involved in cervical carcinogenesis, i.e., cell encoded cell adhesion molecule 1 (CADM1), T-lymphocyte maturation-associated protein (MAL), and microRNA-124-2 (miR124-2), is a marker for an advanced transforming HPV infection [14,15,16,17,18,19,20,21,22]. During HPV-induced cervical carcinogenesis, the methylation levels increase with the severity of the underlying cervical disease and are exceptionally high in cervical cancer [23, 24]. In addition, CIN2/3 lesions with a long-standing (≥5 years) HPV infection have a cancer-like methylation profile and many chromosomal abnormalities, in contrast to CIN2/3 lesions with a recently acquired infection (<5 years) [25]. Accordingly, a positive methylation marker status suggests the presence of a so-called advanced transforming CIN, in need of treatment [21]. In order to further explore our search for biomarkers which lend support to a standardized diagnosis of CIN and identify lesions in need for treatment, we evaluated expression of the HPV E4 protein and hypermethylation of CADM1, MAL, and miR124-2 genes in cervical lesions stratified by both classical CIN grading and the Ki-67 and p16ink4a immunoscore values.
Materials and methods
Study population
We selected 115 formalin-fixed paraffin-embedded cervical biopsy and large loop excision of the transformation zone specimens from the files of the Pathology Department (VU University Medical Center, Amsterdam, The Netherlands) as previously described [6]. The specimens were anonymously processed and selection was guided by initial diagnosis of disease (22 nondysplastic lesions, 22 CIN1, 27 CIN2, 22 CIN3, and 22 squamous cell carcinoma). Ethical approval was waived according to the regulations in The Netherlands [26].
Immunohistochemistry
Serial sections of 3 μm were cut from all tissue blocks. To ensure the presence of the same lesion in all specimens, the first and last sections were stained for H&E (sandwich technique). In between sections were immunostained with mouse monoclonal antibodies (mAb) against Ki-67 antigen (Clone MIB-1, DAKO, Denmark) or p16ink4a antigen (Clone E6H4™, CINtec®, Roche, Switzerland) by the automated IHC Ventana staining machine (Ventana Medical Systems, Roche, USA), or with the validated mAb panHPVE4 (further referred to as “E4”, mAb FH1.1, produced in the laboratory of J. Doorbar, previously described by van Baars et al., reactive against high-risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67, and 70) [2, 12]. For E4 staining, slides were deparaffinized in xylene and rehydrated in a descending alcohol series. FH1.1 primary antibody was applied at a concentration of 1:500 after 30-min microwave pretreatment in Tris/EDTA buffer (10 mM Tris/1 mM EDTA, pH 9.0) and incubated overnight at 4 °C. Application of the primary antibody was followed by incubation with BrightVision plus Poly-HRP anti-mouse IgG (ImmunoLogic, The Netherlands), diaminobenzidine as a chromagen and Hematoxylin nuclear counterstaining.
Molecular testing
DNA was isolated from formalin-fixed paraffin-embedded specimens by a proteinase K procedure and analyzed for the presence of high-risk HPV DNA by GP5+/6+ PCR-EIA (DDL, Rijswijk, the Netherlands) [27]. In addition the isolated DNA was subjected to bisulfite treatment using the EZ DNA Methylation Kit (Zymo Research, California, USA) for DNA methylation analysis [14, 15, 24, 28]. For the analysis of hypermethylation of host-cell genes CADM1, MAL, and miR124-2, a quantitative methylation specific PCR (PreCursor-M assay, Self-screen B.V., Amsterdam, The Netherlands) was performed on the ABI 7500 Fast Real-Time PCR System (Applied biosystems, California, USA) according to the manufacturer’s instructions and as previously described [23]. The housekeeping gene β-actin was tested as methylation-independent reference. All samples had a quantification cycle (Cq) value for β-actin < 33 to assure DNA quality and successful bisulfite conversion. Methylation marker results were expressed in Cq ratios calculated by the following formula: 2 [Cq (β-actin) − Cq (methylation marker)] × 100.
CIN grading
Three expert gyneco-pathologists (M.B., D.J., and M.vd.S.) independently rendered CIN grades (either no dysplasia, CIN1, CIN2, CIN3, or squamous cell carcinoma), using current CIN grading criteria [1]. They then provided Ki-67 (score: 0–3) and p16ink4a (score: 0–3) scores independently and without taking morphologic features into account, as recently described [6]. For Ki-67 scoring, nuclear Ki-67 staining in squamous cells was scored positive. Staining predominantly found in the basal layer was considered normal and scored as 0. Predominant staining of the lower one-third, two-third, or more than two-third of the epithelium was scored as 1, 2, or 3, respectively. For p16ink4a scoring, diffuse or “block” staining of the cytoplasm or nucleus of squamous epithelial cells was considered positive. Absence of p16ink4a positivity or a few scattered positive cells (patchy staining) were scored as 0. Diffuse, low intensity staining limited to the lower one-third of the epithelium was scored as 1, continuous positivity in the lower two-third was scored as 2, and diffuse staining involving the full thickness of the epithelium was scored as 3. These Ki-67 and p16ink4a scores were combined cumulatively into the immunoscore value (ranging from 0 to 6). Immunostains of the E4 protein were subsequently scored as either negative, focally positive (restricted to the upper quarter of the epithelium) or extensively positive (upper one-third of the epithelium or more) [2]. Majority consensus scores of CIN grades, immunoscores, and E4 were used, and based on agreement of two out of three pathologists. If there was no majority, consensus was reached in a panel-discussion with a fourth pathologist (C.M.). The pathologists were blinded to the results from HPV and methylation marker testing.
Statistical analysis
Hypermethylation status (negative or positive) for the CADM1, MAL, and miR124-2 marker panel was determined in all samples and considered positive if the Cq ratios of at least one of the individual methylation markers was above the threshold for positivity. This threshold was calculated per methylation marker by the following formula: (average of Cq ratios of specimens without dysplasia) + (2.58 × [standard deviation of Cq ratios of specimens without dysplasia]).
Expression of E4 (negative, focal, or extensive) and methylation marker status were independently determined for each specimen and stratified by CIN grade and Ki-67 and p16ink4a immunoscore.
In addition, the correlation between methylation marker status on one hand, and CIN grades or Ki-67 and p16ink4a immunoscore groups on the other, was evaluated by Fisher’s exact statistical analysis with p < 0.05 considered significant. The immunoscore groups were defined as previously described, based on immunoscore values 0–3, 4–5, and 6 [6]. The difference in methylation marker status between CIN3 and ≤CIN1, or between lesions defined by immunoscore 6 and immunoscore 0–3, were again determined by Fisher’s exact statistical analysis. Cervical carcinomas were excluded in the correlation analyses.
Calculations were performed in Microsoft Excel (2010), SPSS (V.22), STATA (V14.1) and Graphpad (V7).
Results
CIN grade, immunoscore, and HPV status
HPV results in relation to consensus histology and immunoscore data are shown in Table 1. Consensus CIN grading revealed the following scores: no dysplasia in 35 specimens, CIN1 in 19 specimens, CIN2 in 17 specimens, CIN3 in 22 specimens, and squamous cell carcinoma in 22 specimens. Consensus Ki-67 and p16ink4a immunoscoring revealed the following scores: immunoscore 0–3 in 50 specimens, immunoscore 4 and 5 in 13 specimens and immunoscore 6 in 30 specimens. In all carcinomas, an immunoscore of 6 was found.
E4 expression
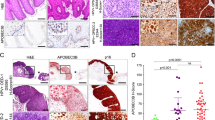
As shown in Table 2, E4 expression was absent in specimens with no dysplasia and was present in 7 (37%) CIN1, 7 (41%) CIN2, 7 (32%) CIN3, and 5 (23%) squamous cell carcinomas. E4 expression was only present in lesions which tested HPV positive (Table 3). Extensive expression of E4 was highest in CIN1 with a slight decrease via CIN2 and CIN3 to absent in cancer. Extensive E4 expression was found in 6 (32%) of the CIN1 lesions, in 3 (18%) of the CIN2 lesions, in 3 (14%) of the CIN3 lesions, and in none of the carcinomas. Focal expression was found in 1 (5%) of the CIN1 lesions, in 4 (24%) of CIN2 lesions, in 4 (18%) of CIN3 lesions, and 5 (23%) of the carcinomas. The E4 expression observed in some of the CIN3 and carcinomas was mostly present in the upper, less atypical parts of the lesion, and only sometimes E4 expression was seen in severely atypical cells (Fig. 1e). Cervical lesions classified by the immunoscore grading system showed a comparable expression pattern (Table 2).
Examples of E4 protein expression and hypermethylation of CADM1, MAL, or miR124-2 according to cervical intraepithelial neoplasia (CIN) grading and immunoscore grading. a No dysplasia; basal Ki-67 positivity and no p16 ; no E4 positivity and no hypermethylation for genes CADM1, MAL, or miR124-2. b CIN1; parabasal Ki-67 positivity, patchy p16 positivity; extensive E4 expression and no hypermethylation for genes CADM1, MAL, or miR124-2. c CIN2; full thickness Ki-67 positivity and p16 positivity; extensive E4 expression and no hypermethylation for genes CADM1, MAL, or miR124-2. d CIN3; full thickness Ki-67 positivity and p16 positivity; no E4 expression and positive hypermethylation for genes CADM1, MAL, or miR124-2. e CIN3; full thickness Ki-67 positivity and p16 positivity; extensive E4 expression and negative hypermethylation for genes CADM1, MAL, or miR124-2
Methylation marker status
Hypermethylation levels of CADM1, MAL, and miR124-2 markers increased with the severity of cervical disease. Four (11%) specimens without dysplasia were borderline positive (i.e., Cq ratios just above the threshold for marker positivity). Two (11%) of the CIN1 lesions, 3 (18%) of the CIN2 lesions, 13 (59%) of the CIN3 lesions, and all 22 (100%) of the squamous cell carcinomas showed hypermethylation of the CADM1, MAL, and miR124-2 marker panel (Table 2). Furthermore, the proportion of markers testing hypermethylation positive (i.e., one, two, or three) increased with the severity of cervical lesions (Fig. 2). This varied from none out of three markers positive in the majority of nondysplastic lesions to mainly all three markers positive in cervical carcinomas. Hypermethylation marker status was significantly correlated to classical CIN grades (Fisher’s exact p < 0.001), as well as to cervical lesions defined by immunoscore groups (Fisher’s exact p = 0.001). Methylation marker status significantly differed between CIN3 and ≤CIN1 (Fisher’s exact p < 0.001), as well as between lesions defined by immunoscore 6 compared to lesions defined by immunoscore 0–3 (Fisher’s exact p < 0.001).
Correlated expression of biomarkers
The correlation of expression of E4 and methylation markers in cervical lesions is shown in Table 4 and illustrated by examples in Fig. 1. In general, expression of E4 protein and hypermethylation of the CADM1, MAL, and miR124-2 marker panel were inversely related, confirming that cervical lesions with signs of both productive HPV infection and HPV transformation are relatively rare but still found in one CIN3 (immunoscore 6).
In more detail, high-grade CIN lesions (either defined as CIN2/3 or by immunoscore 4–6) with extensive E4 expression tested negative for the methylation panel CADM1, MAL, and miR124-2 in all but one case (proportion of test positives: for CIN2/3: n = 1/6, 17%, and immunoscore 4–6: n = 1/8, 13%). Also, in all low-grade lesions (≤CIN1 or ≤immunoscore 3) with extensive E4 expression, no methylation marker positivity was found. On the other hand, high-grade lesions with no extensive E4 expression showed hypermethylation more often (for CIN2/3: n = 15/33, 46%; for immunoscore 4–6: n = 16/35, 46%).
Discussion
In this study, we determined expression patterns for both the E4 protein, reflecting the onset of a productive HPV infection, and hypermethylation status of host-cell genes CADM1, MAL, and miR124-2, associated with transformation by HPV infection, in a series of cervical precursor lesions with increasing grade of severity and cancer [21, 29, 30]. E4 expression correlated with low hypermethylation of the host-cell genes, irrespective of lesion grade. Extensive E4 expression was found most frequent in CIN1 lesions and decreased with increasing CIN grade to be absent in cervical carcinomas, especially when analyzed in HPV-positive lesions only. E4 was not found in HPV negative lesions. On the contrary, positivity rate of hypermethylation of CADM1, MAL, and miR124-2 genes was very low in CIN1 and lesions with Ki-67 and p16ink4a immunoscore 0–3, and increased with the severity of CIN, detecting all carcinomas. Also the proportion of hypermethylated CADM1, MAL, or miR124-2 genes increased with increasing CIN grade toward cancer. Nonetheless, extensive expression of E4 was present in a percentage of all categories of CIN defined by both CIN and Ki-67 and p16ink4a immunoscore grading, indicating that some lesions with transforming features (CIN2/3 or Ki-67 and p16ink4a immunoscore 4–6) may still support late events in the papillomavirus life cycle. In general, however, E4 expression correlated with low-hypermethylation levels of genes CADM1, MAL, or miR124-2, irrespective of lesion grade.
In previous literature, Griffin et al. [12, 31] state that the inverse pattern of transformation and E4 in cervical disease may facilitate the detection and monitoring of low-grade lesions, and their transition to higher-grade disease. Our data confirm that extensive E4 expression is found in low-grade lesions, but also in the upper epithelial layers of some high-grade lesions (illustrated by example in Fig. 1e). Previous studies have shown that E4 expression is much more common in CIN2 than CIN3, suggesting that CIN2 is very heterogeneous and includes predominantly productive lesions as well as more transformed lesions. The expression of E4 in some higher-grade lesions including CIN3 illustrates the importance of Ki-67 and p16ink4a and hypermethylation of host-cell genes to complement E4. Increased E6/E7 deregulation, as present in Ki-67 and p16ink4a positive lesions, can potentially result in suppression of E4, and also lead to upregulation of DNMT1, causing increased hypermethylation and transformation of cells [13, 32]. Accordingly, we found very low hypermethylation levels in extensive E4 positive lesions, which suggests that E4 is a surrogate of methylation absence. Collectively, our results further support the use of E4 and methylation analysis of host-cell genes in combination with other biomarkers, as additional tools to define more accurately early (E4 positive) and advanced CIN lesions (Ki-67, p16ink4a, and hypermethylation positive). This concept is presented in Fig. 3.
Proposed conceptual scheme of cervical carcinogenesis and the extensive expression of E4 and hypermethylation within different stages of cervical disease. MM levels of hypermethylation and number of genes testing positive, increased with the severity of disease; IS immunoscore; CIN cervical intraepithelial neoplasia; HPV human papillomavirus
A limitation of our study is that clinically validated thresholds to determine CADM1, MAL, and miR124-2 marker’s positivity in DNA isolates from cervical formalin-fixed paraffin-embedded material are not yet available, as this marker panel has been validated before in isolates from cervical scrape material [33]. To resolve this, we determined thresholds for positivity based on the Cq ratios of the nondysplastic tissue specimens. In future studies, these thresholds specific for DNA isolates from formalin-fixed paraffin-embedded specimens need to be further validated. Furthermore, we used a protein K procedure on whole-tissue sections for DNA isolation from formalin-fixed paraffin-embedded material. This technique is feasible and yields a high quantity of DNA. However, we cannot exclude target dilution because sometimes only a small part of the tissue section consisted of dysplastic cells. Laser capture microdissection might help to solve this problem [34].
We previously showed that the Ki-67 and p16ink4a immunoscore grading system has a higher accuracy and higher reproducibility compared to classical CIN grading, particularly for the diagnosis of CIN3 (treatment) and CIN1 (no treatment) thereby narrowing the number of the heterogeneous CIN2 group of lesions. Our results involving E4 expression and hypermethylation further support significance of the use of Ki-67 and p16ink4a staining for accurate CIN grading. The significant differences we found between lesions defined by an immunoscore of 6 (treatment) and an immunoscore of 0–3 (no treatment) in methylation marker status and the low frequency of hypermethylation in lesions with extensive E4 expression further substantiate the use of the immunoscore grading system in order to determine clinical management in women diagnosed with CIN.
In particular, the heterogeneous group of CIN2 with immunoscores ≥4, wherein management might vary, can be divided by expression of E4, indicating the onset of a productive infection, into lesions with a low short-term progression risk to cancer on one hand, and by hypermethylation status, indicating an advanced transforming infection, into lesions with a high short-term progression risk on the other hand. In clinical practice, extensive expression of E4 might be used to advocate a wait-and-see policy, whereas positivity for hypermethylation status might trigger the decision to treat. Such a biomarker-based approach for classification of CIN has the potential to prevent under- and overtreatment in women diagnosed with cervical lesions. Although promising, larger studies are needed to validate our findings before we can make definitive clinical recommendations. Finally it should be realized that even in the hands of an experienced colposcopist, the sensitivity of the tissue biopsy for CIN2 or CIN3 does not exceed 50–60% [35, 36], making the use of a more objective, biomarker-based CIN grading system even more necessary.
In conclusion, our results on HPV E4 protein expression and hypermethylation status of CADM1, MAL, and miR124-2 genes in cervical lesions defined either by CIN grading or by Ki-67 and p16ink4a immunoscore grading argue for the use of these biomarkers as adjunctive tools to substantiate an accurate CIN diagnosis. Some extensive E4 expression was found in CIN lesions defined by Ki-67 and p16ink4a immunoscore groups 4–6, however, these lesions showed very low hypermethylation of CADM1, MAL, and miR124-2 genes and the E4 expression was often present in the upper, less atypical parts of a lesion. This further substantiates the gradual transition from early CIN lesions, characterized by extensive E4 expression, to advanced transforming lesions, characterized by extensive hypermethylation of CADM1, MAL, and miR124-2 genes. The use of E4 expression and hypermethylation status in addition to Ki-67 and p16ink4a expression is likely to result in a more accurate approach to identify CIN lesions in need of treatment. Larger studies to confirm the value of defining the biomarker patterns defined here, and establish their value in decisions about treatment of potential precancers in clinical practice are needed.
References
Wright TW, Ronnett BM, Kurman RJ, Ferenczy A, Kurman RJ, Ellenson LH. et al. Blaustein’s Pathology of the Female Genital Tract. Bosten, MA: Springer; 2011. 2011:193–252.
van Baars R, Griffin H, Wu Z, et al. Investigating diagnostic problems of CIN1 and CIN2 associated with high-risk HPV by combining the novel molecular biomarker PanHPVE4 with P16INK4a. Am J Surg Pathol. 2015;39:1518–28.
Ostor AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993;12:186–92.
Baak JP, Kruse AJ, Robboy SJ, et al. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol. 2006;59:1017–28.
Castle PE, Schiffman M, Wheeler CM, Solomon D. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol. 2009;113:18–25.
van Zummeren M, Leeman A, Kremer WW, et al. A three-tiered score for Ki-67 and p16ink4a improves accuracy and reproducibility of grading CIN lesions. J Clin Pathol. In press.
Darragh TM, Colgan TJ, Cox JT, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205–42.
Herrington CS. The terminology of pre-invasive cervical lesions in the UK cervical screening programme. Cytopathology. 2015;26:346–50.
Heatley MK. How should we grade CIN? Histopathology. 2002;40:377–90.
Kyrgiou M, Mitra A, Arbyn M, et al. Fertility and early pregnancy outcomes after treatment for cervical intraepithelial neoplasia: systematic review and meta-analysis. Br Med J. 2014;349:g6192.
Doorbar J, Quint W, Banks L, et al. The biology and life cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70.
Griffin H, Soneji Y, Van Baars R, et al. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod Pathol. 2015;28:977–93.
Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98.
Overmeer RM, Henken FE, Snijders PJ, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008;215:388–97.
Overmeer RM, Henken FE, Bierkens M, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;219:327–36.
Wilting SM, van Boerdonk RA, Henken FE, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167.
Overmeer RM, Louwers JA, Meijer CJ, et al. Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int J Cancer. 2011;129:2218–25.
De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila). 2014;7:1251–7.
Verlaat W, Snijders PJ, Novianti PW, et al. Genome-wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical pre-cancer and cancer. Clin Cancer Res. 2017;23:3813–22.
Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–9.
Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405.
Luttmer R, De Strooper LM, Steenbergen RD, et al. Management of high-risk HPV-positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn. 2016;16:961–74.
De Strooper LM, van Zummeren M, Steenbergen RD, et al. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;67:1067–71.
Van Zummeren M, Kremer WW, Van Aardt MC, et al. Selection of women at risk for cervical cancer in an HIV-infected SOUTH AFRICAN population. AIDS. 2017;31:1945–53.
Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;133:1293–9.
Federation of Biomedical Scientific Societies. Human Tissue and Medical Research: Code of Conduct for responsible use. https://www.federa.org/sites/default/files/images/print_version_code_of_conduct_english.pdf (2011).
van den Brule AJ, Pol R, Fransen-Daalmeijer N, et al. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J Clin Microbiol. 2002;40:779–87.
Sie D, Snijders PJ, Meijer GA, et al. Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell Oncol (Dordr). 2014;37:353–61.
Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25(Suppl 1):2–23.
Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Prim. 2016;2:16086.
Griffin H, Wu Z, Marnane R, et al. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS ONE. 2012;7:e49974.
Burgers WA, Blanchon L, Pradhan S, et al. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–5.
Snellenberg S, De Strooper LM, Hesselink AT, et al. Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape. BMCw Cancer. 2012;12:551–9.
van Baars R, van der Marel J, Snijders PJ, et al. CADM1 and MAL methylation status in cervical scrapes is representative of the most severe underlying lesion in women with multiple cervical biopsies. Int J Cancer. 2016;138:463–71.
van der Marel J, van Baars R, Rodriguez A, et al. The increased detection of cervical intraepithelial neoplasia when using a second biopsy at colposcopy. Gynecol Oncol. 2014;135:201–7.
Wentzensen N, Walker JL, Gold MA, et al. Multiple biopsies and detection of cervical cancer precursors at colposcopy. J Clin Oncol. 2015;33:83–9.
Acknowledgments
We gratefully acknowledge the research staff and technicians of the Department of Pathology VUmc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
(1) D.A.M.H., P.J.F.S., R.D.M.S., and C.J.L.M.M. are minority shareholders of self-screen B.V., a spin-off company of VUmc; (2) self-screen B.V. holds patents related to the work (i.e., hrHPV test and methylation markers for cervical screening); (3) D.A.M.H. has been on the speaker's bureau of Qiagen and serves occasionally on the scientific advisory boards of Pfizer and Bristol-Meyers Squibb; (4) P.J.F.S. has been on the speakers bureau of Roche diagnostics, Gen-Probe, Abbott, Qiagen and Seegene and has been a consultant for Crucell B.V.; (5) J.B. received consultancy fees from Roche, GlaxoSmithKline, and Merck and received travel support from DDL. All fees were collected by his employer; (6) C.J.L.M.M. served occasionally on the scientific advisory board (expert meeting) of Qiagen and SPMSD/Merck, and has by occasion been consultant for Qiagen; he has been co- investigator on a Sanofi Pasteur/MSD sponsored HPV vaccination trial in men of which his institute received research funding; (7) C.J.L.M.M. has small number of shares of Qiagen, is minority shareholder of Self-Screen bv of which he is part-time director since sept 2017. He was minority shareholder of Diassay B.V. until April 2016; (8) M.V.Z., W.W.K., A.L., M.C.G.B., D.J., J.D., and G.G.K. declare that they have no conflict of interest.
Additional information
Deceased: Peter J. F. Snijders
Rights and permissions
About this article
Cite this article
Zummeren, M.v., Kremer, W.W., Leeman, A. et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124-2 genes in cervical cancer and precursor lesions. Mod Pathol 31, 1842–1850 (2018). https://doi.org/10.1038/s41379-018-0101-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0101-z
This article is cited by
-
MAL expression downregulation through suppressive H3K27me3 marks at the promoter in HPV16-related cervical cancers is prognostically relevant and manifested by the interplay of novel MAL antisense long noncoding RNA AC103563.8, E7 oncoprotein and EZH2
Clinical Epigenetics (2024)
-
MiR-214 inhibits apoptosis in thyroid epithelial follicular cells induced by amiodarone through the FASL/MAPK pathway
Molecular & Cellular Toxicology (2022)
-
A whole-slide image grading benchmark and tissue classification for cervical cancer precursor lesions with inter-observer variability
Medical & Biological Engineering & Computing (2021)
-
Characterization of cervical biopsies of women with HIV and HPV co-infection using p16ink4a, ki-67 and HPV E4 immunohistochemistry and DNA methylation
Modern Pathology (2020)
-
Histomorphologic assessment and distribution of high-risk human papillomavirus (HPV) types in cervical high-grade squamous intraepithelial lesions with unusual histomorphologic features
Virchows Archiv (2020)