Abstract
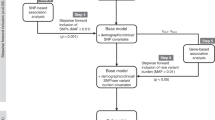
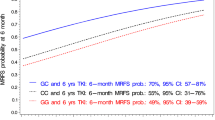
The antileukaemic drug 6-mercaptopurine is converted into thioguanine nucleotides (TGN) and incorporated into DNA (DNA-TG), the active end metabolite. In a series of genome-wide association studies, we analysed time-weighted means (wm) of erythrocyte concentrations of TGN (Ery-TGN) and DNA-TG in 1009 patients undergoing maintenance therapy for acute lymphoblastic leukaemia (ALL). In discovery analyses (454 patients), the propensity for DNA-TG incorporation (wmDNA-TG/wmEry-TGN ratio) was significantly associated with three intronic SNPs in NT5C2 (top hit: rs72846714; P = 2.09 × 10−10, minor allele frequency 15%). In validation analyses (555 patients), this association remained significant during both early and late maintenance therapy (P = 8.4 × 10−6 and 1.3 × 10−3, respectively). The association was mostly driven by differences in wmEry-TGN, but in regression analyses adjusted for wmEry-TGN (P < 0.0001), rs72846714-A genotype was also associated with a higher wmDNA-TG (P = 0.029). Targeted sequencing of NT5C2 did not identify any missense variants associated with rs72846714 or wmEry-TGN/wmDNA-TG. rs72846714 was not associated with relapse risk, but in a separate cohort of 180 children with relapsed ALL, rs72846714-A genotype was associated with increased occurrence of relapse-specific NT5C2 gain-of-function mutations that reduce cytosol TGN levels (P = 0.03). These observations highlight the impact of both germline and acquired mutations in drug metabolism and disease trajectory.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Elion GB. The purine path to chemotherapy. Biosci Rep. 1989;9:509–29.
De Beaumais TA, Jacqz-Aigrain E. Pharmacogenetic determinants of mercaptopurine disposition in children with acute lymphoblastic leukemia. Eur J Clin Pharmacol. 2012;68:1233–42.
Balis FM, Holcenberg JS, Poplack DG, Ge J, Sather HN, Murphy RF, et al. Pharmacokinetics and pharmacodynamics of oral methotrexate and mercaptopurine in children with lower risk acute lymphoblastic leukemia: a Joint Children’s Cancer Group and Pediatric Oncology Branch Study. Blood. 1998;92:3569–77.
Karran P. Thiopurines, DNA damage, DNA repair and therapy-related cancer. Br Med Bull. 2006;79–80:153–70.
Schmiegelow K, Nielsen SN, Frandsen TL, Nersting J. Mercaptopurine/methotrexate maintenance therapy of childhood acute lymphoblastic leukemia: clinical facts and fiction. J Pediatr Hematol Oncol. 2014;36:503–17.
Evensen NA, Madhusoodhan PP, Meyer J, Saliba J, Chowdhury A, Araten DJ, et al. MSH6 haploinsufficiency at relapse contributes to the development of thiopurine resistance in pediatric B-lymphoblastic leukemia. Haematologica. 2018;2017:176362.
Nielsen SN, Grell K, Nersting J, Frandsen TL, Hjalgrim LL, Schmiegelow K. Measures of 6-mercaptopurine and methotrexate maintenance therapy intensity in childhood acute lymphoblastic leukemia. Cancer Chemother Pharmacol . 2016;78:983–94.
Nielsen SN, Grell K, Nersting J, Abrahamsson J, Lund B, Kanerva J, et al. DNA-thioguanine nucleotide concentration and relapse-free survival during maintenance therapy of childhood acute lymphoblastic leukaemia (NOPHO ALL2008): a prospective substudy of a phase 3 trial. Lancet Oncol. 2017;2045:1–10.
Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–8.
Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui C-H, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–91.
Toft N, Birgens H, Abrahamsson J, Bernell P, Griškevičius L, Hallböök H, et al. Risk group assignment differs for children and adults 1-45 yr with acute lymphoblastic leukemia treated by the NOPHO ALL-2008 protocol. Eur J Haematol. 2013;90:404–12.
Frandsen TL, Heyman M, Abrahamsson J, Vettenranta K, Åsberg A, Vaitkeviciene G, et al. Complying with the European Clinical Trials directive while surviving the administrative pressure - an alternative approach to toxicity registration in a cancer trial. Eur J Cancer. 2014;50:251–9.
Toft N, Birgens H, Abrahamsson J, Griškevičius L, Hallböök H, Heyman M. et al. Results of NOPHO ALL2008 treatment for patients aged 1–45 years with acute lymphoblastic leukemia. Leukemia. 2018;32:606–15.
Tulstrup M, Frandsen TL, Abrahamsson J, Lund B, Vettenranta K, Jonsson OG, et al. Individualized 6-mercaptopurine increments in consolidation treatment of childhood acute lymphoblastic leukemia: a NOPHO randomized controlled trial. Eur J Haematol. 2017;93:1–8.
Jacobsen JH, Schmiegelow K, Nersting J. Liquid chromatography-tandem mass spectrometry quantification of 6-thioguanine in DNA using endogenous guanine as internal standard. J Chromatogr B Anal Technol Biomed Life Sci. 2012;881–2:115–8.
Shipkova M, Armstrong VW, Wieland E, Oellerich M. Differences in nucleotide hydrolysis contribute to the differences between erythrocyte 6-thioguanine nucleotide concentrations determined by two widely used methods. Clin Chem. 2003;49:260–8.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Core Team R. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015.
Yang JJ, Landier W, Yang W, Liu C, Hageman L, Cheng, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33:1235–42.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:1–14.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:930–4.
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Zabala W, Cruz R, Barreiro-de Acosta M, Chaparro M, Panes J, Echarri A, et al. New genetic associations in thiopurine-related bone marrow toxicity among inflammatory bowel disease patients. Pharmacogenomics. 2013;14:631–40.
Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43.
Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:2869–79.
Spychala J, Madrid-Marina V, Fox IH. High K(m) soluble 5’-nucleotidase from human placenta. Properties and allosteric regulation by IMP and ATP. J Biol Chem. 1988;263:18759–65.
Brouwer C, Vogels-Mentink TM, Keizer-Garritsen JJ, Trijbels FJM, Bökkerink JPM, Hoogerbrugge PM, et al. Role of 5′-nucleotidase in thiopurine metabolism: enzyme kinetic profile and association with thio-GMP levels in patients with acute lymphoblastic leukemia during 6-mercaptopurine treatment. Clin Chim Acta. 2005;361:95–103.
Gazziola C, Ferraro P, Moras M, Reichard P, Bianchi V. Cytosolic high K(m) 5′-nucleotidase and 5′(3′)-deoxyribonucleotidase in substrate cycles involved in nucleotide metabolism. J Biol Chem. 2001;276:6185–90.
Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–71.
Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–4.
Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90:1699–701.
Emadi A, Karp JE. The clinically relevant pharmacogenomic changes in acute myelogenous leukemia. Pharmacogenomics. 2012;13:1257–69.
Mitra AK, Crews KR, Pounds S, Cao X, Feldberg T, Ghodke Y, et al. Genetic variants in cytosolic 5’-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J Pharmacol Exp Ther. 2011;339:9–23.
Gamazon ER, Lamba JK, Pounds S, Stark AL, Wheeler HE, Cao X, et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood. 2013;121:4366–76.
Jordheim LP, Nguyen-Dumont T, Thomas X, Dumontet C, Tavtigian SV. Differential allelic expression in leukoblast from patients with acute myeloid leukemia suggests genetic regulation of CDA, DCK, NT5C2, NT5C3, and TP53. Drug Metab Dispos. 2008;36:2419–23.
Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sánchez-Martín M, Madubata CJ, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553:511–4.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604.
Acknowledgements
This work was supported by The Danish Cancer Society, The Danish Childhood Cancer Foundation, The Swedish Childhood Cancer Foundation, The Nordic Cancer Union, The Otto Christensen Foundation, University Hospital Rigshospitalet and The Novo Nordic Foundation.
Author contributions
MT compiled the data and handled bioinformatics analyses with MG, SNN, BOW and RG. MT and KG handled statistical analyses. MT drafted the manuscript. PSW, OGJ, BL, AH-S, JA, GV, KP, NT, MH, EH, SL, LG and MP developed the study protocol and coordinated the national blood sample and data collection for each country. JW and WLC assisted with data and analyses from the TARGET database. ZZ designed the probes for the sequencing panel. MDD designed and performed the DNA library preparation for targeted gene sequencing. JN supervised all pharmacological analyses. KS initiated and supervised the study, was the principal investigator for this study and the NOPHO ALL2008 protocol, and had responsibility for the final submission for publication. All authors approved the final manuscript
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tulstrup, M., Grosjean, M., Nielsen, S.N. et al. NT5C2 germline variants alter thiopurine metabolism and are associated with acquired NT5C2 relapse mutations in childhood acute lymphoblastic leukaemia. Leukemia 32, 2527–2535 (2018). https://doi.org/10.1038/s41375-018-0245-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0245-3
This article is cited by
-
Maintenance therapy for acute lymphoblastic leukemia: basic science and clinical translations
Leukemia (2022)
-
NT5DC2 promotes tumor cell proliferation by stabilizing EGFR in hepatocellular carcinoma
Cell Death & Disease (2020)
-
Somatic and germline genomics in paediatric acute lymphoblastic leukaemia
Nature Reviews Clinical Oncology (2019)