Abstract
MODY 5 and 6 have been shown to be low-penetrant MODYs. As the genetic background of unknown MODY is assumed to be similar, a new analytical strategy is applied here to elucidate genetic predispositions to unknown MODY. We examined to find whether there are major MODY gene loci remaining to be identified using SNP linkage analysis in Japanese. Whole-exome sequencing was performed with seven families with typical MODY. Candidates for novel MODY genes were examined combined with in silico network analysis. Some peaks were found only in either parametric or non-parametric analysis; however, none of these peaks showed a LOD score greater than 3.7, which is approved to be the significance threshold of evidence for linkage. Exome sequencing revealed that three mutated genes were common among 3 families and 42 mutated genes were common in two families. Only one of these genes, MYO5A, having rare amino acid mutations p.R849Q and p.V1601G, was involved in the biological network of known MODY genes through the intermediary of the INS. Although only one promising candidate gene, MYO5A, was identified, no novel, high penetrant MODY genes might remain to be found in Japanese MODY.
Similar content being viewed by others
Introduction
Of the type 2 diabetes mellitus (T2DM) susceptibility single-nucleotide polymorphisms (SNPs) identified before the use of genome-wide association studies (GWAS) [1], the pathogenic mechanisms are well understood partly based on the results of studies in monogenic diabetes [2]. It is therefore strategically reasonable to apply information of pathophysiology obtained from rare monogenic DM to that from common T2DM.
One such example of monogenic (Mendelian) diabetes is maturity-onset diabetes of the young (MODY), which is classically characterized by autosomal dominant inheritance, onset before the age of 25 years in at least one family member and partly preserved pancreatic beta cell function [3]. Since MODY has similar clinical features as T2DM in the Japanese population and both diseases are characterized by non-obesity and impaired insulin secretion, MODY is highly relevant as a model diabetes. Six major genes have been found to cause MODY sub-forms in both Asian and Caucasian populations; it is now possible to tailor and optimize treatment based on genetic diagnosis, especially for MODY3 [4]. MODY is inherited in a Mendelian fashion and the number of MODY patients in Japan is at most ~500,000; however, 70% of the responsible genes are still unknown in Japanese MODY families.
There has also recently been little progress in identifying MODY genes in Caucasians. This is due to the fact that there are few multigeneration families that remain to be analyzed and that, unlike in Japan, close subclassification based on phenotype is difficult in terms of healthcare costs. Japanese patients with MODY are relatively hypoinsulinemic due to their intrinsically lower capacity of insulin secretion compared to that of Caucasians and this contributes to the development of diabetes even in low-penetrant MODYs [5, 6]. It should therefore be advantageous to closely examine Japanese MODY candidates for genetic mutations. Although MODY has been classified as a single gene defective diabetes, it has been clarified that the development of MODY5 and MODY6 is influenced by other modifying factors including ethnicity, specific genetic background and/or intrauterine environment, leading to the concept that MODYs, especially low-penetrant MODYs, have characteristics of common type 2 diabetes having multifactorial origin.
In the present study, we first examined whether or not there are major monogenic MODY gene loci in Japanese that remain to be identified using genome-wide SNP linkage analysis. We then applied whole-exome sequencing followed by in silico network analysis. If novel MODY genetic causality is not a single gene mutation (monogenic), there is the potential for an overlapping of low-penetrant mutations (oligogenic), identification of which requires extraction of MODY candidate genes in “islet function network” units.
Material and methods
Subjects
We have so far collected a total of 422 MODY probands who were negative for mutations in MODY1–6 and 10 from 155 medical institutions throughout Japan having diabetes suspected to be MODY (male/female, 196/226; age of diagnosis, 17.7 ± 8.4 years; BMI, 23.8 ± 5.1 kg/m2; insulin (Ins) /oral hypoglycemic agents (OHA) /Diet/GLP-1RA/NA, 204/69/71/3/75; and family history (yes/no/NA, 302/50/70). The screening criteria used were onset-age of 35 years or less, anti-GAD antibody negativity and no severe obesity (BMI<30 kg/m2). Out of the provided MODY families, the responsible gene for which is unknown, we selected pedigrees having the genome of at least three members. These consist of 95 samples, 31 of which are probands and the rest are family samples. The probands were recruited from medical institutions throughout Japan with diabetes whose disease was suspected to be MODY (male/female 12/19; age of onset, 14.3 ± 7.3 years; BMI, 22.0 ± 5.0 kg/m2). Eighteen of the probands were treated with Ins, 7 with OHA, 4 with diet only, and 2 with no information. The 64 family members consist of 41 diabetic, 18 non-diabetic, and 5 subjects of unknown affected status. Family history was not included so as not to miss sporadic or low-penetrant cases [5, 6]. The study protocol was approved by the Institutional Review Board of Gifu University (No. 25-153). Written, informed consent was obtained from all participants. When a candidate missense mutation was detected, whether or not it was a rare polymorphism was determined by direct sequencing of 567 unrelated non-diabetic (control) elderly subjects (male/female 226/341; age, 67.4 ± 6.0 years; BMI, 23.0 ± 2.9 kg/m2; HbA1c, 5.4 ± 0.4% (NGSP) or 36 ± 2.6 mmol/mol (IFCC)) [7].
Linkage analysis by SNP mapping
We performed genome-wide linkage analysis to detect new MODY gene loci with two different SNP datasets, Affymetrix 6.0 array with about 900 K SNPs for 95 individuals from 31 families and VCF files of exome sequencing from 32 individuals from seven families described later.
Genotype calls from SNP chip data were converted by using an application of Affymetrix genotyping arrays and Affymetrix Power Tools. VCF genotype files were obtained from exome data. The linkage mapping input files were converted from BRLMM genotype files of SNP chip and VCF genotype files of exome data by LINKDATAGEN with data from HapMap Phase III Japanese population [8]. The genotype was subjected to a multipoint linkage analysis to detect association with the disease status in all families. LOD scores were calculated using the program MERLIN 1.1.232 [9].
We selected SNPs to meet the criteria of call rate >0.9, HWE in control >0.001, Maximum confidence score 0.05, and Intermarker distance 80–120 kb. We then performed both parametric and non-parametric analyses under the model, that is Parametric analysis: AD model, Disease frequency of 0.005, Phenocopy of 0.005, Penetrance of 0.95, and unknown or unaffected status for T2DM and Non-parametric analysis: Linear model, S-pairs, and Equal contribution of each family.
Exome sequencing and data analyses
Exome sequencing was performed for 32 individuals with typical MODY from 7 families (male/female 2/5; age of onset, 16.3 ± 7.9 years; BMI, 20.5 ± 3.4 kg/m2). We selected pedigrees having the genome of at least three members. Actually, all families but one has more than four genome samples. BMI values of family members of only pedigree 8 are >25 and <30. All other families have BMI of less than 25. Considering our previous cases of patients with low-penetrant MODY5 and MODY6, MODY-X (unknown MODY) might develop to overt diabetes after 25 years of age. We therefore selected criteria including onset-age of 35 years or less. DNA samples (3 μg) were subjected to exome capture using the SureSelect Human All Exon kit version 4.0 (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions and were used for solution hybridization; the enriched target DNA fragment was subjected to sequencing using Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA). Paired-end 100-bp reads were mapped to the reference genome (GRCh37) using Burrows-Wheeler Aligner (BWA) v.0.7.9 [10]. BWA-generated SAM files were converted to BAM format, then sorted and indexed using SAMtools v.0.1.18. 16 [11]. The files obtained in BAM format were analyzed using GATK v.2.7 best practice guidelines [12]. In brief, BAM files were first subjected to indel realignment, base quality score recalibration, and variant calling with the GATK HaplotypeCaller to obtain the potential variants in the VCF file. These variants were annotated using ANNOVAR [13]. Depth-based CNV calls were analyzed by CNVnator [14]. For gene annotation, we used the RefSeq gene database (build hg19) while variant annotation was based on dbSNP (dbSNP 147), 2049 Japanese data in Integrative Japanese Genome Variation (iJGVD; https://ijgvd.megabank.tohoku.ac.jp/about/), 1000 Genomes Project database and Exome Aggregation Consortium (ExAC) version 0.3.1 (http://exac.broadinstitute.org). Functional effect of the variants was evaluated by SIFT [15], PolyPhen2 [16], Protein Variation Effect Analyzer (PROVEAN) [17] and Combined Annotation Dependent Depletion (CADD) [18] for scoring the deleteriousness of coding variants for clinical validity. The probability of being loss-of-function (LoF) intolerant (pLI) score was based on data from the ExAC, providing exome variants from 60,706 unrelated individuals [19]. The pLI score was used to measure gene-based intolerance.
All variants detected from exome sequencing data were further analyzed by performing three filtering steps based on different criteria: in the first filtering step, we selected variants of missense, nonsense, splice-site single-nucleotide variants (SNV), and insertion or deletions (indels). The second filtering step was based on the frequency in Japanese and world-wide population. Variants with a frequency <0.025% in the iJGVD and 1000 genomes were filtered as MODY candidates. Finally, heterozygous variants co-segregated in the family were selected for candidate MODY causal variants. Functional effect of all candidate variants were scored and categorized for clinical validity, as damaging for SIFT, and deleterious for PolyPhen2 and PROVEAN. After these filtering steps, candidate variants were confirmed for all family members by Sanger sequencing on the 3130xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). Variants of HNF4A, GCK, HNF1A, PDX1, HNF1B, NEUROD1, KLF11, CEL, PAX4, INS, BLK, ABCC8, KCNJ11, and APPL1 have previously been identified in MODY. The candidacy of known MODY genes in the variant list of exome sequencing results was confirmed in the first instance.
Functional enrichment analysis (in silico network analysis)
Network analysis performed in this study was performed by GeneMANIA program (http://www.genemania.org) [20]. This program finds other genes that are related to a set of input genes using a very large set of functional association data such as protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. The details of the pathway were examined carefully on the Pathway Commons website (http://www.pathwaycommons.org/pc/sif_interaction_rules.do).
Results
Mapping of new MODY loci
The Manhattan view of LOD scores identified by genome-wide linkage analysis of our MODY probands and family samples are shown in Fig. 1. Four peaks with over 2.0 LOD score from non-parametric analysis were found in chr2 (2.1), chr4 (2.09), chr13 (2.06), and chr19 (2.01). In addition, 11 of the other 19 autosomal chromosomes had weak LOD scores (>1.5). However, none of peaks showed LOD score more than 3.7, which is approved to be the threshold of evidence for linkage in genome-wide linkage analysis. Therefore, it is highly unlikely that a major MODY gene is shared among the families; conversely, locus heterogeneity, each family having different causality, is likely to explain the result of linkage analysis. To break through the difficulty of identifying new MODY genes, the exome sequencing method was applied to seven families having typical MODY. The linkage analysis for the 32 individuals from the seven families expressed different patterns of LOD score; higher LOD scores (>2.5) were detected in five chromosomes, chr8 (2.89), chr12 (2.83), chr14 (2.69), chr15 (2.68), and chr20 (2.52) (Supplementary Fig. S1). We then focused on candidate genes with deleterious rare amino acid substitutions that were shared among more than two families.
Exome sequencing and data analyses
Exome sequencing was performed with 32 individuals from 7 families. The average coverage depth was 146.4×, with 97.1% of target bases covered by at least five reads. This supports a high level of confidence in the variant calling.
After filtering for the candidate variants by function (missense, nonsense, splice-site SNVs and indels) and frequency (<0.025% in iJGVD and 1000 genomes), 145, 62, 1003, 374, 241, 31, and 52 variants were co-segregated in Family 8, Family 12, Family 15, Family 25, Family ZB, Family HG and Family HS, respectively. Next, filtering of functional effect (categorized as damaging for SIFT and deleterious for PolyPhen2 and PROVEAN), 35, 12, 233, 83, 48, 5, and 7 were identified as damaging or deleterious from Family 8, Family 12, Family 15, Family 25, Family ZB, Family HG and Family HS, respectively. (Supplementary Fig. S2, Supplementary Fig. S3 and Supplementary Table S1). All possible CNVs were not co-segregated with affection status in each family.
Mutated genes shared among pedigrees
Seven families were analyzed by exome sequencing and co-segregated candidate genes ranging from 9 to 236 were detected in these families. The number of genes, 39, 12, 236, 87, 52, 9, and 17, that could be mutated with the rare variants were identified from Family 8, Family 12, Family 15, Family 25, Family ZB, Family HG, and Family HS, respectively. Three mutated genes were common among three families and 42 mutated genes were common in two families. The variant list of 45 candidate genes was sorted by the region with higher LOD Score in chr8, chr12, chr14, chr15, and chr20. Only one gene (MYO5A) with rare amino acid substitutions, c.G2546A:p.R849Q in Family 12 and c.T4802G:p.V1601G in Family 15, was located on the chr15 region identified by linkage analysis. To further evaluate the 45 candidate genes identified, we used the probability of LoF intolerance (pLI). Eleven mutated genes had a high pLI score (pLI > 0.99), identifying an extremely LoF intolerant set of genes (Table 1).
Variants in MYO5A
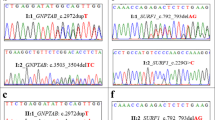
The MYO5A variants p.R849Q in Family 12 and p.V1601G in Family 15 were considered candidate novel MODY gene's variants. The PROVEAN score of p.R849Q and p.V1601G were −3.21 and −6.42, respectively. Highly important, CADD score of p.R849Q and p.V1601G were 7.795 and 5.86, respectively. The CADD scores for the pathogenicity ware greater than the median for known pathogenic variants (3.498). We sequenced two exons of MYO5A to determine whether or not the two mutations identified in the present study were rare polymorphisms in control subjects and we could not find them. (Fig. 2, Table 2, Fig. 3). The patients with the mutations showed severe diabetic retinopathy and nephropathy irrespective of the disease duration. They also showed mild obesity and developed diabetes within <10 years (Fig. 3, Family 12; II-2, Family 15; I-2, II-1).
Location of two mutations identified in the present study and functional domains of Myosin Va protein. Full-length Myosin Va protein contains a core of the myosin structure (MYSc) defined as motor activity (GO:0003774) and ATP binding (GO:0005524), an IQ calmodulin-binding motif (GO:0005515), and a dilute (DIL) domain with unknown function. The R849Q mutation from Family 12 is located in the domain of the IQ calmodulin-binding motif and the p.V1601G (p.V1574G) is in the DIL domain
Genotypes of the MYO5A mutations identified in two families. Families with the R849Q and V1601G mutation of MYO5A. The annotation indicates sample name and genotype. The proband is indicated by the arrow. Since we concentrated to get all candidate variants shared with definite MODY patients who met our criteria for MODY, filled in black, we did not use unconfirmed MODY samples, filled in gray from each pedigree. The white circle and square indicate normal glucose tolerant subjects by OGTT. The individual I-1 in Family 15 was not analyzed
Functional enrichment analysis
Most of the fourteen, known MODY genes are found in the same pathways, insulin processing and secretion. We expected that novel MODY genes might be in the biological network of known MODY genes related to insulin secretion. Physical interaction and pathway analyses among the fourteen, known MODY genes and 45 candidate genes were performed to search for biological interaction by using GeneMANIA. Only one pathway network cluster including ten genes, HNF4A, GCK, HNF1A, PDX1, NEUROD1, PAX4, INS, ABCC8, KCNJ11, and MYO5A was constructed with an additional four genes as intermediary. MYO5A was listed in our expression genes’ catalog of human islet tumor and also rat normal islets, but not in RINm5F, which has no capacity of insulin secretion [21, 22]. Surprisingly, only one gene, MYO5A, was involved in the biological network of known MODY genes through the intermediary of the insulin gene, INS (Fig. 4) The MYO5A gene is found in both the pathway of insulin processing, and secretion due to translocation of GLUT4 to the plasma membrane (Table 1).
Functional enrichment analysis for novel candidate MODY genes. Network connections involving the fourteen known MODY genes and 45 candidate genes. The orange circles are candidate genes from the exome analysis; the green circles are from the known MODY genes; and the black circles are genes associated with both of the genes. The blue lines show Pathway connections between related genes via biological datasets and the red lines show Physical Interactions. The line width reflects the weight of networks among gene interactions as closely as possible with genes on the input list and as little as possible with genes not on the list. The thick lines show high evidence for inclusion on the network
Discussion
Although 90 or more genes responsible for T2DM have been identified so far by GWAS, they are nevertheless not better onset predictors than environmental factors such as gender, age, and body weight [23]. Against this background, the present study focuses on the analysis of “monogenic” MODY, which is often inherited in an autosomal dominant fashion and for which the effect of mutation of the genes responsible is clear. While DNA sequencing of the entire genome in many individuals is still unfeasible and very expensive, targeted sequencing of the coding regions of all ~20,000 human genes by next-generation sequencer is practically possible. Thus, exome sequencing is well justified as an efficient strategy to search for mutations underlying Mendelian disorders such as MODY, especially since the great majority of disease-causing mutations disrupt a protein-coding sequence. In fact, protein-coding exons are estimated to harbor about 85% of the mutations with large impact on disease-related traits [24].
However, there has recently been little progress in identifying novel MODY genes; only MODY 7–14 genes have been reported with family-based studies in the eighteen years since the MODY6 gene was identified [25]. This is partly due to the fact that there are few multigeneration families that remain to be analyzed both in the Caucasian and Japanese population. Another possible reason for not finding novel MODY genes is an incomplete penetrance of the phenotype of a mutation of mild effect and/or interactions with disease modifying factors. In addition, it is undeniable that families have different causal genes. Therefore, even with the latest techniques of exome sequencing analysis, it is likely that identification of novel MODY genes would be difficult by sequencing with only a small number of family members.
MODY1–6 accounts for about 30% of all Japanese MODY, and 70% are still unknown. We have reported that in low-penetrant MODYs such as MODY5 and 6, the complexities of genetic background among races can significantly affect the occurrence of the disease [5, 6]. Japanese readily develop overt diabetes as their capacity of insulin secretion is intrinsically low; it is therefore advantageous to explore for novel MODY genes, especially low-penetrant MODY genes, in Japanese patients. We have received requests for analysis of responsible genes from medical institutions throughout Japan. We selected criteria including onset-age of 35 years or less, autoimmune antibody negativity and no severe obesity. Family history such as three consecutive generations of diabetes was not included so as not to miss sporadic or low-penetrant cases [5, 6].
In the past several years, genetic diagnosis has been based on PCR, Sanger sequencing and MLPA, even though only 20% of patients have their disease diagnosed. Therefore, in this study we integrated diverse strategies such as linkage analysis, exome sequencing analysis, and in silico network analysis with knowledge database.
As a first strategy, genome-wide linkage analysis was applied to identify high penetrant and frequent MODY genes to explain many of the cases. However, the results of linkage analysis in the present study, in which different and diffused LOD scores were found, indicates that no major MODY genes are left to discover in Japanese, suggesting that novel MODY genes could be specific to only a few families.
As a breakthrough strategy for discovering new MODY genes, exome sequencing was performed for seven families with typical MODY including 32 individuals. As expected from the results of linkage analysis, MODY candidate genes seem to have locus heterogeneity and to be specific for each family. Since the variants on the shared, mutated genes could be higher penetrant and good candidates for the disease, the number of candidate genes was narrowed down to 45 genes by selection according to the number of overlapping families. Finally, physical interaction and pathway analyses among the fourteen, known MODY genes and 45 candidate genes were performed, resulting in the identification of only one gene, MYO5A, that is involved in the biological network of known MODY genes through the intermediary of INS.
Only one gene, MYO5A, also was acceptable as a novel candidate gene by functional enrichment analysis and was connected to INS coding insulin via the pathway of insulin processing/synthesis/secretion. MYO5A encoding Myosin Va protein plays a role in translocation of the insulin-containing secretory granules across the cytosol to the inner surface of the plasma membrane [26]. Myosin V consists of two heavy chains containing the myosin head and a neck with six IQ motifs [27]. The two MYO5A variants are located in domain IQ and the DIL domain. IQ, the calmodulin-binding motif, is a functional regulation region as the motility of myosin V is inhibited by calcium. Whereas the DIL domain in globular tail regions of myosin V has no known function, the importance of the tail region of myosin V is specified for cargo binding with Rab27, which is known to be associated with a number of myosin V cargo vesicles such as GLUT4 [28].
Griscelli syndromes are known to be caused by mutations in the Rab27 gene [29] and mutations in the myosin V motor protein gene [30, 31]. Griscelli syndrome type 1 is caused by homozygous mutations of the MYO5A gene, and is characterized by hypopigmentation with frequent pyogenic infection, enlargement of the liver and spleen, low blood neutrophil level, low blood platelet level, and immunodeficiency. Diabetes-related traits have not yet been examined adequately in conditions of both homozygous and heterozygous mutations. In melanocytes, melanosomes (vesicles containing the pigment melanin) are transported on microtubules. They are then bound by Rab27A, which recruits synaptotagmin-like protein lacking C2 domains A (SLAC2A) and myosin Va (MYO5A). This complex then transfers the melanosomes from the microtubules to actin filaments. This transfer is necessary for the transport of melanosomes from the perinuclear area to the cell periphery. The loss of any one of these proteins interrupts melanosome transport and results in hypopigmentation.
Regarding diabetes-related traits, fasting insulin and insulin resistance, in silico replication in the MAGIC study showed weak but significant association (adjusted P-value of 0.0097) for rs34602777 in the MYO5A gene [32]. Using Agilent Human Whole-Genome arrays, genes that were predicted targets of miR-26b, miR-30b, and miR-145 were upregulated in insulin resistant subjects. These were examined, resulting in the identification of ADAM22, MYO5A, LOX, and GM2A as predicted gene targets of the microRNAs [33]. The patients with the mutations showed severe diabetic complications, especially nephropathy irrespective of the duration of diabetes. They showed mild obesity at an early stage and developed diabetes in a short period possibly due to the relatively earlier reduction of insulin secretion. Further accumulation of cases with mutations of MYO5A is necessary for elucidation of the role of MYO5A in the pathophysiology of diabetes.
New findings that will be obtained from our analysis of MODY in the present study can be expected to provide clues for understanding the mechanism of impaired insulin secretion and/or insulin resistance, and will contribute insight into the onset mechanism of “polygenic types” of T2DM and the development of novel therapies, although a functional analysis will be necessary to elucidate the mechanism by which dysfunction of MYO5A affects insulin secretion and action.
However, there are many likely pathogenic variants found in exome sequencing as genes of uncertain significance (GUS). The evidence of GUS rare variants is not sufficient to explain a biological function as a MODY mechanism; the findings will require follow-up studies to obtain a sufficient evidence level required for clinical support by confirmation of the significance through identification of causative mutations in other MODY families.
There are a few limitations of the study: the biological effect of R859Q and V1601G in MYO5A were not examined. We excerpted from biological study of function of domain IQ and DIL, where two variants are located. We mentioned the function of each domain and discussed the possibility of biological mal-effect as disease causality. However, functional studies for MYO5A domain DIL are especially difficult due to unknown function of DIL domain. Furthermore, we did not read the whole genome including enhancer regions and also did not examine the change of methylation.
In conclusion, although only one promising candidate gene, MYO5A, was identified in the present study, no major genetic factor in Japanese MODY of unknown origin was found and the possibility that locus heterogeneity of low-penetrant genes is causal in the rest of the MODY families became clear.
ACCESSION NUMBERS: All sequence data associated with this project has been submitted to Japanese Genotype–Phenotype Archive (JGA Study ID: JGAS00000000072, JGA Dataset ID: JGAD00000000072, http://trace.ddbj.nig.ac.jp/jga/index_e.html).
References
Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47.
Hattersley AT, Patel KA. Precision diabetes: learning from monogenic diabetes. Diabetologia. 2017;60:769–77.
Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–80.
Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362:1275–81.
Horikawa Y, Enya M, Fushimi N, Fushimi Y, Takeda J. Screening of diabetes of youth for HNF-1 mutations: clinical phenotype of HNF1B-MODY and HNF1A-MODY in Japanese. Diabet Med. 2014;31:721–7.
Horikawa Y, Enya M, Mabe H, Fukushima K, Takubo N, Ohashi M, et al. NEUROD1-deficient diabetes (MODY6): Identification of the first cases in Japanese and the clinical features. Pediatr Diabetes. 2018;19:236–42. https://doi.org/10.1111/pedi.12553
Enya M, Horikawa Y, Iizuka K, et al. Association of genetic variants of the incretin-related genes with quantitative traits and occurrence of type 2 diabetes in Japanese. Mol Genet Metab Rep. 2014;1:350–61.
Smith KR, Bromhead CJ, Hildebrand MS, Shearer AE, Lockhart PJ, Najmabadi H, et al. Reducing the exome search space for mendelian diseases using genetic linkage analysis of exome genotypes. Genome Biol. 2011;2:R85.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense variants. Nat Methods. 2010;7:248–9.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Exome Aggregation Consortium:. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4.
Wang H, Horikawa Y, Jin L, Narita T, Yamada S, Shihara N, et al. Gene expression profile in rat pancreatic islet and RINm5F cells. J Mol Endocrinol. 2005;35:1–12.
Jin L, Wang H, Narita T, Kikuno R, Ohara O, Shihara N, et al. Expression profile of mRNAs from human pancreatic islet tumors. J Mol Endocrinol. 2003;31:519–28.
McCarthy MI, Hattersley AT. Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes. 2008;57:2889–98.
Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13 https://doi.org/10.1186/gm13
Johansson S, Irgens H, Chudasama KK, Molnes J, Aerts J, Roque FS, et al. Exome sequencing and genetic testing for MODY. PLoS One. 2012;7:e38050.
Rutter GA, Hill EV. Insulin vesicle release: walk, kiss, pause… then run. Physiology (Bethesda). 2006;21:189–96.
Cheney RE, O’Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, et al. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23.
Fan F, Matsunaga K, Wang H, Ishizaki R, Kobayashi E, Kiyonari H, et al. Exophilin-8 assembles secretory granules for exocytosis in the actin cortex via interaction with RIM-BP2 and myosin-VIIa. eLife. 2017;6:e26174 https://doi.org/10.7554/eLife.26174
Menasche G, Ho CH, Sanal O, Feldmann J, Tezcan I, Ersoy F, et al. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). J Clin Invest. 2003;112:450–6.
Anikster Y, Huizing M, Anderson PD, Fitzpatrick DL, Klar A, Gross-Kieselstein E, et al. Evidence that Griscelli syndrome with neurological involvement is caused by mutations in RAB27A, not MYO5A. Am J Hum Genet. 2002;71:407–14.
Pastural E, Barrat FJ, Dufourcq-Lagelouse R, Certain S, Sanal O, Jabado N, et al. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat Genet. 1997;16:289–92.
Chen G, Bentley A, Adeyemo A, Shriner D, Zhou J, Doumatey A, et al. Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum Mol Genet. 2012;21:4530–6.
Kirby TJ, Walton RG, Finlin B, Zhu B, Unal R, Rasouli N, et al. Integrative mRNA-microRNA analyses reveal novel interactions related to insulin sensitivity in human adipose tissue. Physiol Genomics. 2016;48:145–53.
Acknowledgements
The authors are very grateful to the patients and their families for their participation in the study. We thank K Yokoyama, J Kawada, and H Tsuchida for technical assistance.
Author contributions
YH, KH, ST, SS, II, and JT contributed to the study design. YH, KH, ME, HI, and YS performed the genetic experiments and analyzed and interpreted the genetic data. YH, KH, and JT wrote the manuscript. All coauthors read and approved the manuscript.
Funding
This work was supported by a Health and Labor Science Research Grant for Research on Rare and Intractable Diseases from the Japanese Ministry of Health, Labor and Welfare, a Grant-in-Aid for Scientific Research from the Japanese Ministry of Science, Education, Sports, Culture and Technology, and a Strategic International Research Cooperative Program Grant from Japan Science and Technology Agency.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
10038_2018_449_MOESM2_ESM.tif
Supplementary Fig. S2 LOD scores of linkage analysis of 32 individuals from seven MODY families with typical modalities of MODY(TIF 28683 kb)
Rights and permissions
About this article
Cite this article
Horikawa, Y., Hosomichi, K., Enya, M. et al. No novel, high penetrant gene might remain to be found in Japanese patients with unknown MODY. J Hum Genet 63, 821–829 (2018). https://doi.org/10.1038/s10038-018-0449-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0449-4
This article is cited by
-
Monogenic diabetes
Diabetology International (2024)