Abstract
Cornelia de Lange syndrome (CdLS) is a rare multisystem disorder with specific dysmorphic features. Pathogenic genetic variants encoding cohesion complex subunits and interacting proteins (e.g., NIPBL, SMC1A, SMC3, HDAC8, and RAD21) are the major causes of CdLS. However, there are many clinically diagnosed cases of CdLS without pathogenic variants in these genes. To identify further genetic causes of CdLS, we performed whole-exome sequencing in 57 CdLS families, systematically evaluating both single nucleotides variants (SNVs) and copy number variations (CNVs). We identified pathogenic genetic changes in 36 out of 57 (63.2 %) families, including 32 SNVs and four CNVs. Two known CdLS genes, NIPBL and SMC1A, were mutated in 23 and two cases, respectively. Among the remaining 32 individuals, four genes (ANKRD11, EP300, KMT2A, and SETD5) each harbored a pathogenic variant in a single individual. These variants are known to be involved in CdLS-like. Furthermore, pathogenic CNVs were detected in NIPBL, MED13L, and EHMT1, along with pathogenic SNVs in ZMYND11, MED13L, and PHIP. These three latter genes were involved in diseases other than CdLS and CdLS-like. Systematic clinical evaluation of all patients using a recently proposed clinical scoring system showed that ZMYND11, MED13L, and PHIP abnormality may cause CdLS or CdLS-like.
Similar content being viewed by others
Introduction
Cornelia de Lange syndrome (CdLS, MIM #122470, #300590, #610759, #614701, #300882) is a rare neurodevelopmental disorder characterized by dysmorphic features, prenatal onset growth restriction, hirsutism, upper limb reduction defects (which range from subtle phalangeal abnormalities to oligodactyly), developmental delay, and intellectual disability [1]. Prevalence of CdLS has been estimated at 1/10,000–1/30,000 of live births [2]. In addition to these cardinal phenotypes, patients show cardiac anomalies, gastroesophageal reflux, seizures, and behavioral problems [3]. A combination of signs and symptoms define the classic CdLS phenotype, which is easily recognized from birth by experienced pediatricians and clinical geneticists. However, CdLS is a genetically heterogeneous disorder presenting with extensive phenotypic variability from mild to severe, and with different degrees of facial and limb abnormalities. In addition, CdLS clinically overlaps with several other diseases, including Bohring-Optiz syndrome, CHOPS syndrome, and Fryns syndrome [4, 5]. Such heterogeneity makes it difficult to clearly distinguish CdLS from other clinically overlapping diseases. Recently, an international consensus group provided clinical criteria for CdLS [6]. This criteria uses a scoring system comprised of cardinal and suggestive features.
To date, pathogenic variants in at least 15 genes are known to cause CdLS [7,8,9,10]. In this regard, cohesin complex or its functionally related genes (e.g., nipped B-like protein [NIPBL], structural maintenance of chromosome 1A [SMC1A], SMC3, histone deacetylase 8 [HDAC8], and RAD21 cohesin complex component [RAD21]) have been implicated. Approximately 60% of CdLS patients harbor various NIPBL variants [1]. Cohesin is a multisubunit protein complex consisting of four core proteins: SMC1, SMC3, RAD21, and stromal antigen (STAG) [6]. Chromatin loading of cohesion is regulated by NIPBL [11]. The cohesin complex plays a significant role in mediating sister chromatid cohesion, DNA double-strand break repair, transcriptional regulation, and chromatin organization. Abnormalities of cohesion complex and its related genes in humans are known as cohesinopathy [12]. In addition, variants in AFF4, ANKRD11, ARID1B, BRD4, EP300, ESPL1, KMT2A, PDGFRB, SETD5, and TAF6 also cause a CdLS-like phenotype. [7,8,9, 13,14,15]
In this study, we investigated 57 clinically suspected CdLS individuals by whole-exome sequencing (WES). Genetic findings, including single nucleotide variants (SNVs) and copy number variations (CNVs), together with clinical features obtained using recent clinical criteria are presented and discussed.
Methods
Subjects
In this study, 57 patients were recruited from 57 families, consisting of 56 Brazilian and one Japanese patients. Most of the Brazilian patients were referred by the Brazilian Association of Cornelia de Lange Syndrome (CdLS Brazil) and had the clinical diagnosis suspected by pediatricians and/or geneticists from all over the country based on distinctive features, such as synophrys, arched eyebrows, long philtrum, upper limb abnormalities, and hirsutism. For comparison of clinical manifestations within our cohort and genotype–phenotype correlations, clinical details (including atypical symptoms) were retrospectively reviewed based on recent clinical criteria reported by Kline et al. [6]. Clinical information was obtained from all 57 patients (Table S1). Peripheral blood leukocytes were collected from patients and their parents after obtaining informed consent. Parental samples were available except for five families (Families 6, 7, 10, 22, and 30). This study was approved by the Institutional Review Boards of Yokohama City University, Faculty of Medicine, and University of Sao Paulo, Faculty of Medicine.
Whole-exome sequencing
Genomic DNA was extracted from whole-blood sample using QuickGene-610L (Fujifilm, Tokyo, Japan) according to the manufacturer’s protocol. Genomic DNA was sheared using a S220 Focused-ultrasonicator (Covaris, Woburn, MA, USA) and captured using the SureSelect Human All Exon V6 Kit (Agilent Technologies, Santa Clara, CA, USA). Paired-end libraries were sequenced on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) with 101-bp paired-end reads. Quality-controlled reads were aligned to the human reference genome (UCSC hg19, NCBI build 37.1) using NOVOALIGN (http://www.novocraft.com/products/novoalign/). After removal of polymerase chain reaction (PCR) duplications using Picard (http://broadinstitute.github.io/picard/), variants were called using Genome Analysis Tool Kit (GATK) (https://software.broadinstitute.org/gatk/index.php). Called variants were annotated using ANNOVAR (http://annovar.openbioinformatics.org/en/latest/). Exonic and intronic variants within 30 bp from exon–intron boundaries were examined. Synonymous variants and variants with minor allele frequencies ≧0.01 in our in-house exome database of 575 Japanese individuals or control population databases (including the Exome Aggregation Consortium Browser population (ExAC) [http://exac.broadinstitute.org/] and National Heart, Lung, and Blood Institute (NHLBI) exome variant server [http://evs.gs.washington.edu/EVS/]) were removed. Missense variants were evaluated using Sorting Intolerant From Tolerant (SIFT) (http://sift.jcvi.org/), Polymorphism Phenotyping v2 (Polyphen-2) (http://genetics.bwh.harvard.edu/pph2./), and MutationTaster (http://MutationTaster.org/).
In particular, the focus was on five CdLS genes (NIPBL, SMC1A, SMC3, HDAC8, and RAD21) and 10 CdLS-like genes (AFF4, ANKRD11, ARID1B, BRD4, EP300, ESPL1, KMT2A, PDGFRB, SETD5, and TAF6). Candidate variants were validated by Sanger sequencing. Additionally, de novo occurrences were validated when parental samples were available. Parentage was confirmed by analyzing 12 microsatellite markers with Gene Mapper software v4.1.1 (Life Technologies Inc., Carlsbad, CA, USA). The WES performance is summarized in Supplementary Information (Table S2).
Real-time reverse transcription PCR
To detect aberrant transcripts caused by splice site mutations, reverse transcription PCR (RT-PCR) was performed using total RNA extracted from patient derived lymphoblastoid cell lines. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and reverse-transcribed into cDNA using the Super Script First Strand Synthesis System (Takara, Kyoto, Japan). Resultant cDNA was used as a template for PCR. PCR amplicons were subjected to Sanger sequencing and aberrant transcripts were characterized. For RT-PCR analysis of NIPBL, the forward and reverse primers were: 5′-GAACACTTCAGTTGCTGCAAA-3′ and 5′-CGTTTCCTAGAGGATTCAAAAGC-3′ in Patient 15 with c.3121 + 1G>A, and 5′-TCATCCAGTTCAGTGTGTGC-3′ and 5′-TCTCAATGACCCTGAAGTGC-3′ in Patient 28 with c.7410 + 4A>G.
WES-based CNV analysis
Using WES data, CNVs were analyzed by two algorithms: the eXome Hidden Markov Model (XHMM), and a program based on relative depth of coverage ratio, developed by Nord et al. (Nord program) [16, 17]. For genome-wide screening, XHMM data were first examined in each patient. If causative CNVs were detected using XHMM, altered copy numbers of such regions were further verified using the Nord program. In addition, CNVs at five CdLS genes and 10 CdLS-like genes (see WES section above) were tested by the Nord program.
Quantitative polymerase chain reaction
Candidate CNVs were validated by quantitative polymerase chain reaction (qPCR). Real-time qPCR was performed to examine genomic DNA copy number at NIPBL, C5orf42, MED13L, SMARCA2, FREM1, and EHMT1 target loci. QuantiFast SYBER Green PCR kit (Qiagen) was used for real-time quantification with amplification monitored on a Rotor-Gene Q real-time PCR cycler (Qiagen). Relative ratios of genomic DNA copy number were calculated using the standard curve method with Rotor-Gene 6000 Series Software 1.7 (Qiagen) by normalizing with autosomal internal control loci (STXBP1 and/or FBN1) and also compared to an unrelated control individual. Information of all primers is available on request.
Results
Flowchart of this study
A flowchart of this study is shown in Fig. 1. Because of the genetic and clinical heterogeneity of CdLS, we directly employed WES to effectively screen pathogenic variants in patients with clinically suspected CdLS. To detect variants in CdLS, CdLS-like, or other possible genes, all 57 patients were analyzed based on autosomal dominant (de novo), autosomal recessive, and X-linked modes of inheritance. Based on American College of Medical Genetics and Genomics (ACMG) guidelines [18], we identified 29 pathogenic or likely pathogenic SNVs in two CdLS genes (NIPBL and SMC1A) and four CdLS-like genes (ANKRD11, EP300, KMT2A, and SETD5) (Fig. 1). WES-based CNV analysis in 28 SNV-negative patients detected pathogenic CNVs in four patients (4/57 [7.0%]), involving NIPBL, MED13L, EHMT1, and 9q deletion (Fig. 1). The remaining 24 cases had neither pathogenic SNVs nor CNVs. Consequently, these cases were subjected to trio-based analysis, except for two cases whose parental samples were unavailable. We detected three pathogenic variants in genes associated with diseases other than CdLS and CdLS-like: ZMYND11, MED13L, and PHIP. Altogether, if all abnormalities were included, we identified pathogenic or likely pathogenic variants in 36 out of 57 cases (63.2%) (Fig. 1). Thirty-one of 36 variants occurred de novo, unless biological parental samples were unavailable. One variant was inherited from a mosaic mother (Patient 53). Twenty-three of 32 pathogenic SNVs were novel (Table 1).
Flowchart of this study. All the 57 patients with clinically suspected CdLS were analyzed by whole-exome sequencing (WES). Twenty-nine patients had pathogenic single nucleotide variants (SNVs) in two CdLS genes (NIPBL and SMC1A) and four CdLS-like genes (ANKRD11, EP300, KMT2A, and SETD5). WES-based copy number variation (CNV) analysis in patients with no causative SNVs identified pathogenic CNVs in four patients. The remaining 24 patients with neither pathogenic SNVs nor CNVs were subjected to trio-based analysis, except for two cases whose parental samples were unavailable. Three causative variants were identified in ZMYND11, MED13L, and PHIP. Diagnostic yield was 63.2 % (36/57) when all 32 SNVs (32/57 [56.1%]) and four CNVs (4/57 [7.0%]) were included. A novel candidate variant was detected in NAA50
Pathogenic SNVs in CdLS genes
We detected 22 pathogenic SNVs in NIPBL (22/57 [38.6%]) and two in SMC1A (2/57 [3.5%]) (Table 1 and Fig. 1). Among 22 NIPBL SNVs, 14 were novel. Meanwhile, NM_0133433.3:c.6893G>A, p.Arg2298His was repeatedly detected (Patients 2 and 50). Three splice site variants in NIPBL (NM_0133433.3:c.3121 + 1G>A, c.7410 + 4A>G, and c.5329-15A>G) were detected in Patients 15, 28, and 54, respectively. These variants were previously described, and only c.5329-15A>G was shown to result in abnormal splicing [19,20,21]. The other c.3121 + 1G>A and c.7410 + 4A>G mutations were never examined at cDNA level [19,20,21]. Therefore, by RT-PCR using cDNA derived from lymphoblastoid cells, we confirmed aberrant splicing in both Patient 15, with c.3121 + 1G>A, and Patient 28, with c.7410 + 4A>G (Fig. S1). Regarding the two missense variants in SMC1A, NM_006306.3:c.1152C>G, p.Lys362Asn was novel.
Pathogenic SNVs in CdLS-like genes
We also detected pathogenic variants in four CdLS-like genes: ANKRD11 (2/57 [3.5%]), EP300 (1/57 [1.8%]), KMT2A (1/57 [1.8%]), and SETD5 (1/57 [1.8%]) (Table 1 and Fig. 1), whose pathogenic variants are known to cause KBG syndrome (MIM #148050), Rubinstein–Taybi syndrome 2 (MIM #613684), Wiedemann–Steiner syndrome (MIM #605130), and mental retardation autosomal dominant 23 (MIM #615761), respectively. These disorders all share overlapping clinical features with CdLS. All five variants were novel, occurring de novo except for an EP300 variant, which was due to unavailable parental samples. According to the ACMG guideline, the EP300 variant can be classified as likely pathogenic since it is protein length changing mutation due to in-frame deletion (PM4), it is absent from control (including the ExAC, NHLBI, and gnomAD [https://gnomad.broadinstitute.org/]) (PM2), it is predicted to be deleterious by PROVEAN [http://provean.jcvi.org/seq_submit.php] and CADD [https://cadd.gs.washington.edu/] with a score of 23.6 and 21.1, respectively (PP3), and the phenotype of patient is considered reasonable as Cornelia de Lange syndrome-like (PP4).
Pathogenic CNVs
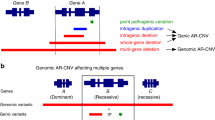
Using the XHMM and Nord program, we detected four pathogenic CNVs in four patients (Table 1 and Fig. 1). These were confirmed by qPCR. Patient 9 has a 94-kb deletion at 5p13.2, encompassing exons 22–47 of NIPBL and the last exon of C5orf42 (Fig. S2,a). Partial deletions of NIPBL have been reported in patients with CdLS, and NIPBL haploinsufficiency is apparently deleterious [22]. Patient 34 has a 4.2-Mb deletion at 12q24.1-q24.23, which contains 40 genes including the entire MED13L gene (Figure S2,b). Patient 51 has a 14.1-Mb deletion at 9p24.3-p22.3, involving 44 genes and an adjacent 571-kb duplication at 9p22.3, altogether encompassing four genes (Fig. S2,c). Patient 52 has a 774-kb deletion at 9q34.3, containing 14 genes including the entire EHMT1 gene (Fig. S2,d).
Variants in genes associated with diseases other than CdLS and CdLS-like
By trio-based analysis, we identified pathogenic or likely pathogenic variants in ZMYND11, MED13L, and PHIP. These variants are involved in other diseases, but never CdLS or CdLS-like.
A novel ZMYND 11 frameshift variant (NM_006624.5:c.1438delG, p.Asp480Thrfs*3) was detected in Patient 53, who had typical CdLS features including left hand oligodactyly (Table 1 and S1, and Fig. 2a–e). Based on apparent double sequences implying low mutant allele peaks in the electropherogram of the mother, maternal mosaicism of this variant was examined (Fig. S3). Deep sequencing of PCR products encompassing the maternal variant confirmed mosaicism (mutant/mutant + wild-type reads = 2835/27596 [10.3%)]), while Patient 53 showed heterozygosity (mutant/mutant + wild-type reads = 12514/27211 [46.0%]) (Table S3). By TA cloning of PCR products spanning the maternal variant, wild-type and mutant alleles were clearly recognized by Sanger sequencing (Fig. S3), yet the mother had no CdLS-like features. ZMYND11 has been reported as a critical gene for 10p15.3 microdeletion syndrome, including neurodevelopmental disorder, characteristic dysmorphic features, and other more frequent symptoms, such as behavioral disturbances, hypotonia, seizures, low birth weight, short stature, genitourinary malformations, and recurrent infections [23].
Clinical photographs of individuals with ZMYND11, MED13L, and PHIP abnormalities. a–e Photos of Patient 53 with a ZMYND11 frameshift mutation. a, b Facial features include microcephaly, synophrys, highly arched eyebrows, long curly eyelashes, low set ears, anteverted nasal nostrilis, long philtrum, thin upper lip, downturned corners of the mouth, and micrognathia. c Note left hand oligodactyly (only one finger). d, e Right hand and bilateral feet. Right hand shows abnormal palmer crease. Feet show no abnormalities. f–h Facial photos of Patient 5 with a MED13L missense mutation at f 3 months and g 18 years. h Broad forehead, synophrys, long curly eyelashes, low set ears, anteverted nasal bridge, and full cheeks are seen at 23 years. i–l Clinical features of Patient 34 with a 4.2-Mb deletion involving MED13L. i Note synophrys, arched eyebrows, upslanting palpebral fissures, long curly eyelashes, low set ears, anteverted nasal bridge, and bulbous nasal tip. j Hirsutism in the back. k, l Bilateral hands and feet. Hands show clinodactyly of the fifth finger. Feet show no abnormalities. m–p Photos of Patient 56 with a PHIP missense mutation. m Facial features include macrocephaly, synophrys, long curly eyelashes, anteverted nasal nostrilis, depressed nasal bridge, and short neck. n Full whole body view with obesity at 11 years (weight, 82.5 kg [>95 percentile]; height, 157.5 cm [>95 percentile]; occipital frontal circumference, 58 cm [>98 percentile]). o, p Hands and feet were normal. q–t Photos of Patient 51 with a 4.1-Mb deletion at 9p24.3-p22.3 adjacent to a 571-kb duplication at 9p22.3. q, r Facial features include synophrys, upslanting palpebral fissures, anteverted nostrilis, and long philtrum. s, t Hands were normal. u–w Phenotype of Patient 52 with a 773.8-kb deletion at 9q34.3. u Facial features include synophrys, long curly eyelashes, and depressed nasal bridge. v, w Hands and feet were normal
A novel MED13L missense mutation, NM_015335.4:c.6485C>A, p.Thr2162Lys was detected in Patient 5 (Table S4). MED13L variants cause distinctive dysmorphic features and mental retardation with or without cardiac defects (MIM #608771), known as MED13L haploinsufficiency syndrome [24]. The missense variant identified here is novel, but another variant at the same nucleotide position was previously identified, which leads to a different amino acid substitution (NM_015335.4:c.6485C>T, p.Thr2162Met) [25]. Of note, we also detected a 4.2-Mb deletion involving MED13L in Patient 34 (Table S4 and Fig. S2b). Further, a novel PHIP missense mutation (NM_017934.7:c.1156G>A, p.Asp386Asn) was detected in Patient 56. PHIP haploinsufficiency causes dysmorphic CdLS-like features, developmental delay, intellectual disability, and obesity [26].
In the remaining 21 undetermined families, NM_025146.4:c.93C>G, p.Tyr31* in NAA50 (encoding N-alpha-acetyltransferase 50) attracted our attention, because it encodes a cohesin complex component (see Discussion). NAA50 variants have not previously been described.
Clinical evaluation of CdLS patients using a new scoring system
Of the 57 patients with CdLS, their clinical features were re-evaluated based on the clinical scoring system reported by Kline et al. [6]. With this scoring system, clinical features of clinically suspected CdLS are classified as cardinal (2 points each if presented) and suggestive (1 point each if presented). Clinical scores ≥ 11, 10 or 9, 8–4, and <4 points, are classified as: classic CdLS, non-classic CdLS, sufficiently suspected to warrant molecular testing for CdLS, and insufficient indication for CdLS molecular testing, respectively. All 57 patients were classified using the above clinical scoring system (Table S1 and Fig. 3). Twenty-five patients were categorized as classic CdLS, 17 patients as non-classic CdLS, and 15 patients as sufficiently suspected to warrant molecular testing for CdLS. No patients were insufficient to indicate molecular testing. The proportion of NIPBL variants was 60% (15/25), 35.3% (6/17), and 13.3% (2/15) in each class, respectively. Ratios of NIPBL variants were compared between two of three classes, with a significant difference recognized only between classic CdLS and sufficiently suspected to warrant molecular testing for CdLS (χ2 test, p < 0.05) (Fig. 3). NIPBL variants in classic CdLS were more frequent than sufficiently suspected to warrant molecular testing for CdLS.
Classification of 57 CdLS patients by clinical score. All patients were classified based on clinical score. Scores of ≥11, 10 or 9, 8–4, and <4 enabled categorization of four classes: classic CdLS, non-classic CdLS, sufficiently suspected to warrant molecular testing for CdLS indicated, and insufficient to indicate molecular testing for CdLS. The 57 patients were classified to classic CdLS (N = 25), non-classic CdLS (N = 17), and sufficiently suspected to warrant molecular testing for CdLS indicated (N = 15). The number of individuals with variants are indicated in rows of genes
Interestingly, Patient 53 with a ZMYND11 frameshift variant showed classic CdLS (15 points) with oligodactyly (Fig. 2a–e). Therefore ZMYND11 could be included as a CdLS or CdLS-like genes, although ZMYND11 variants have not been reported in CdLS. Patients 5 (SNV) and 34 (CNV) with MED13L abnormality showed clinical scores of 8 and 9 points, respectively, and were consequently classified as sufficiently suspected to warrant molecular testing for CdLS and non-classic CdLS (Fig. 2f–h, i–l). Patient 56 with a missense variant in PHIP showed CdLS-like features (6 points), including synophrys, long curly eyelashes, anteverted nostrilis, and depressed nasal bridge, although obesity was retrospectively inconsistent with CdLS (Fig. 2m–p). This clinical information is summarized in Table S1.
Discussion
Using WES, we identified pathogenic variants in 36 out of 57 (63.2%) patients with clinically suspected CdLS. The diagnostic yield was comparatively higher than previous studies (40–60%) as previous studies used panel or Sanger sequencing of only major CdLS genes [27,28,29,30]. Advantages of WES are clearly indicated here as CdLS and CdLS-like patients are genetically and clinically heterogeneous. Using a large clinical exome sequencing cohort, a recent genotype-driven approach of cohesinopathy also emphasized the utility of clinical exome sequencing to provide molecular diagnoses for cohesinopathies with extensive genetic and phenotypic heterogeneity, as well as to detecting mosaic variants in patients [12]. We detected no mosaicism variants in our patients, although it may be difficult to detect extremely low prevalence mosaic variants by WES.
Based on recent clinical scores [6], NIPBL variants are more likely to be found in classic CdLS. Moreover, we detected a ZMYND11 frameshift variant, NM_006624.5:c.1438delG, p.Asp480Thrfs*3 in Patient 53 with classic CdLS. ZMYND11 (also known as BS69) contains a tandem “reader” module of histone modifications, which recognizes and binds histone H3.3 trimethylated at Lys-36 (H3.3K36me3). Subsequently, this recruits histone demethylases, histone deacetylases, and the SWI/SNF chromatin-remodeling complex to reset chromatin to a relatively repressive state and prevent further transcription [31, 32]. Except for ZMYND11, all pathogenic variants in genes for diseases other than CdLS and CdLS-like (MED13L and PHIP) were detected in patients with scores <9.
We found two patients with MED13L abnormality and one patient with a PHIP variant. MED13 is a subunit of the cyclin-dependent kinase 8 (CDK8) module comprised of reversible association of four subunits: cyclin C, CDK8, mediator complex subunit (MED)12/MED12L, and MED13/MED13L. The module binds the mediator complex to regulate its activity. The mediator complex bridges between gene-specific activators bound to regulatory elements and general transcription machinery comprising RNA polymerase II and general transcription factors [33, 34]. PHIP is a H3K4 methylation-binding protein that interacts with chromatin modifications associated with promoters and transcriptional cis-regulatory elements [35]. Interestingly, ZMYND11, MED13L, and PHIP are all core components of transcriptional regulatory pathways. Recently, CdLS and CdLS-like disorders were reported not only as cohesinopathies but also as “transcriptomopathies” [15]. Actually, AFF4, ANKRD11, ARID1B, BRD4, EP300, KMT2A, SETD5, and TAF6 have been found in patients with several clinical features overlapping with CdLS, and are related to epigenetic modification, chromatin remodeling, and transcriptional regulation pathway [8, 15, 36, 37] (Table S5). Interactive networks of 18 genes associated with CdLS and CdLS-like features were analyzed using GeneMANIA (https://genemania.org/), which covers physical interactions, pathways, and shared protein domains (Fig. 4). As expected, genes encoding cohesion complex and its regulatory factors (NIBPL, SMC1A, SMC3, HDAC8, RAD21, and ESPL1) strongly interact with each other. ZMYND11 and PHIP share protein domains with other genes encoding histone modification factors and transcriptional regulation factors. MED13L shares a common pathway with EP300, and is involved in regulation of RNA polymerase II. HIF1A is a hypoxia inducible factor subunit that induces recruitment of CDK8-mediator complex and p300 (encoding EP300) for histone acetyltransferase to stimulate RNA polymerase II elongation [38]. These functional links in three genes (ZMYND11, PHIP, and MED13L) may be related to CdLS-like features.
Schematic presentation of interacting networks of mutated genes in CdLS and CdLS-like. Interactive gene networks of mutated genes with CdLS and CdLS-like features. Three networks are highlighted using GeneMANIA (https://genemania.org/), based on physical interactions (red line), connecting pathways (blue line), and shared protein domains (green line)
Patient 51 has a 14.1-Mb deletion at 9p24.3-p22.3 (involving 44 RefSeq genes) adjacent to a 571-kb duplication at 9p22.3, containing four genes (Fig. S2c). Critical genes of 9p deletion syndrome include DMRT (DMRT1, DMRT2, and DMRT3 cluster) for gonadal dysgenesis from complete sex reversal to milder phenotypes in 46,XY patients [39], FREM1 for craniosynostosis including trigonocephaly [40], and DOCK8, KANK1, SLC1A1, and GLDC for developmental delay and neurological disorders [41]. Trigonocephaly is one of the major features of 9p deletion syndrome, but absent in our patient. Trigonocephaly was previously mapped to a critical 4.7-Mb region at 9p22.2-p23, including FREM1 and CER1 [42]. Interestingly, our patient has a duplication of this critical region, and instead of trigonocephaly, exhibited delayed closure of the anterior fontanelle at 3 years of age Thus, it is conceivable that FREM1 and/or CER1 are potentially dosage sensitive genes related to cranial bone development and closure. In addition, SMARCA2 was included in the deletion region. SMARCA2 is a known causative gene for Nicolaides–Baraitser syndrome (MIM #601358), which shares several CdLS features [43]. To date, 78 variants are registered in the Human Gene Mutation Database (HGMD) V.2019.1, but no truncating variants. SMARCA2 variants are predicted to act in a dominant-negative or gain-of-function manner rather than haploinsufficiency. Indeed, it has been suggested that SMARCA2 might not be a critical gene for 9p deletion syndrome.
Patient 52 has a 773.8-kb deletion at 9q34.3, which contains 14 genes including EHMT1. Intragenic EHMT1 variants or submicroscopic 9q34.3 deletion causes Kleefstra syndrome with distinct facial features, hypotonia, developmental delay, and intellectual disability [44]. EHMT1 encodes a histone H3 Lys-9 methyltransferase and is consequently involved in chromatin remodeling [45]. Similar to patients with CdLS, our patient showed dysmorphic features, including synophrys, long curly eyelashes, and depressed nasal bridge, but no limb abnormalities (Fig. 2u–w). Clinical score was 5 points, suggesting that the patient is likely compatible with Kleefstra syndrome rather than CdLS. Nonetheless, it is sometimes difficult to clearly differentiate these two disorders.
In 21 undetermined families, a de novo nonsense variant (NM_025146.4:c.93C>G, p.Tyr319*) was detected in NAA50 in Patient 19 with classic CdLS features (12 points). The variant was confirmed by Sanger sequencing. This variant was not registered in control population databases (ExAC and gnomAD). According to ExAC, probability of loss-of-function intolerance (pLI) score of 0.88 suggest intolerance to loss-of-function variant. To date, no variants are registered in HGMD V.2019.1. NAA50 encodes a N-terminal acetyltransferase required for chromosome segregation during mitosis. It has been reported that NAA50 is required for sister chromatid cohesion during Drosophila wing development, and most likely regulates correct interaction between the cohesin subunits, RAD21 and SMC3 [46]. These findings support that NAA50 truncation variants may cause the candidate variants of CdLS. Further studies of NAA50 variants in patients with CdLS are necessary.
In conclusion, we have achieved a high genetic diagnosis rate of 63.2% by WES in patients with clinically diagnosed CdLS. Moreover, we have newly detected ZMYND11, MED13L, and PHIP variants potentially linked to CdLS or CdLS-like through abnormality of transcriptional regulation together with NAA50 variant.
References
Nizon M, Henry M, Michot C, Baumann C, Bazin A, Bessieres B, et al. A series of 38 novel germline and somatic mutations of NIPBL in Cornelia de Lange syndrome. Clin Genet 2016;89:584–89.
Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum Mutat 2013;34:1589–96.
Bhuiyan ZA, Klein M, Hammond P, van Haeringen A, Mannens MM, Van Berckelaer-Onnes I, et al. Genotype-phenotype correlations of 39 patients with Cornelia De Lange syndrome: the Dutch experience. J Med Genet 2006;43:568–75.
Bedoukian E, Copenheaver D, Bale S, Deardorff M. Bohring-Opitz syndrome caused by an ASXL1 mutation inherited from a germline mosaic mother. Am J Med Genet Part A 2018;176:1249–52.
McInerney-Leo AM, Harris JE, Gattas M, Peach EE, Sinnott S, Dudding-Byth T, et al. Fryns syndrome associated with recessive mutations in PIGN in two separate families. Hum Mutat 2016;37:695–702.
Kline AD, Moss JF, Selicorni A, Bisgaard AM, Deardorff MA, Gillett PM, et al. Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet 2018;19:649–66.
Parenti I, Teresa-Rodrigo ME, Pozojevic J, Ruiz Gil S, Bader I, Braunholz D, et al. Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum Genet 2017;136:307–20.
Olley G, Ansari M, Bengani H, Grimes GR, Rhodes J, von Kriegsheim A, et al. BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome. Nat Genet 2018;50:329–32.
Ansari M, Poke G, Ferry Q, Williamson K, Aldridge R, Meynert AM, et al. Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J Med Genet 2014;51:659–68.
Yavarna T, Al-Dewik N, Al-Mureikhi M, Ali R, Al-Mesaifri F, Mahmoud L, et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum Genet 2015;134:967–80.
Muto A, Schilling TF. Zebrafish as a Model to Study Cohesin and Cohesinopathies. In: Clifton NJ (editor). Methods in Molecular Biology. New York: Springer Science+Business Media; 2017, pp 177–196.
Yuan B, Neira J, Pehlivan D, Santiago-Sim T, Song X, Rosenfeld J, et al. Clinical exome sequencing reveals locus heterogeneity and phenotypic variability of cohesinopathies. Genet Med: Off J Am Coll Med Genet 2019;3:663–75.
Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet 2015;47:338–44.
Woods SA, Robinson HB, Kohler LJ, Agamanolis D, Sterbenz G, Khalifa M. Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am J Med Genet Part A 2014;164a:251–58.
Yuan B, Pehlivan D, Karaca E, Patel N, Charng WL, Gambin T, et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Investig 2015;125:636–51.
Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 2012;91:597–607.
Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genom 2011;12:184.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med: Off J Am Coll Med Genet 2015;17:405–24.
Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, et al. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet 2007;72:98–108.
Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet 2004;75:610–23.
Teresa-Rodrigo ME, Eckhold J, Puisac B, Pozojevic J, Parenti I, Baquero-Montoya C, et al. Identification and functional characterization of two intronic NIPBL mutations in two patients with Cornelia de Lange syndrome. BioMed Res Intl. 2016;2016:1–8.
Cheng YW, Tan CA, Minor A, Arndt K, Wysinger L, Grange DK, et al. Copy number analysis of NIPBL in a cohort of 510 patients reveals rare copy number variants and a mosaic deletion. Mol Genet Genom Med 2014;2:115–23.
Tumiene B, Ciuladaite Z, Preiksaitiene E, Mameniskiene R, Utkus A, Kucinskas V. Phenotype comparison confirms ZMYND11 as a critical gene for 10p15.3 microdeletion syndrome. J Appl Genet 2017;58:467–74.
Smol T, Petit F, Piton A, Keren B, Sanlaville D, Afenjar A, et al. MED13L-related intellectual disability: involvement of missense variants and delineation of the phenotype. Neurogenetics 2018;19:93–103.
Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med 2017;9:43.
Jansen S, Hoischen A, Coe BP, Carvill GL, Van Esch H, Bosch DGM, et al. A genotype-first approach identifies an intellectual disability-overweight syndrome caused by PHIP haploinsufficiency. Eur J Hum Genet 2018;26:54–63.
Mariani M, Decimi V, Bettini LR, Maitz S, Gervasini C, Masciadri M, et al. Adolescents and adults affected by Cornelia de Lange syndrome: a report of 73 Italian patients. Am J Med Genet Part C, Semin Med Genet 2016;172:206–13.
Marchisio P, Selicorni A, Bianchini S, Milani D, Baggi E, Cerutti M, et al. Audiological findings, genotype and clinical severity score in Cornelia de Lange syndrome. Int J Pediatr Otorhinolaryngol 2014;78:1045–48.
Hei M, Gao X, Wu L. Clinical and genetic study of 20 patients from China with Cornelia de Lange syndrome. BMC Pediatr 2018;18:64.
Mehta D, Vergano SA, Deardorff M, Aggarwal S, Barot A, Johnson DM, et al. Characterization of limb differences in children with Cornelia de Lange syndrome. Am J Med Genet Part C, Semin Med Genet 2016;172:155–62.
Wen H, Li Y, Li H, Shi X. ZMYND11: an H3.3-specific reader of H3K36me3. Cell Cycle 2014;13:2153–54.
Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 2014;508:263–68.
Utami KH, Winata CL, Hillmer AM, Aksoy I, Long HT, Liany H, et al. Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum Mutat 2014;35:1311–20.
Miao YL, Gambini A, Zhang Y, Padilla-Banks E, Jefferson WN, Bernhardt ML, et al. Mediator complex component MED13 regulates zygotic genome activation and is required for postimplantation development in the mouse. Biol Reprod 2018;98:449–64.
Morgan MAJ, Rickels RA, Collings CK, He X, Cao K, Herz HM, et al. A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev 2017;31:2003–14.
Izumi K. Disorders of transcriptional regulation: an emerging category of multiple malformation syndromes. Mol Syndromol. 2016;7:262–73.
Ka M, Kim WY. ANKRD11 associated with intellectual disability and autism regulates dendrite differentiation via the BDNF/TrkB signaling pathway. Neurobiol Dis 2018;111:138–52.
Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013;153:1327–39.
Onesimo R, Orteschi D, Scalzone M, Rossodivita A, Nanni L, Zannoni GF, et al. Chromosome 9p deletion syndrome and sex reversal: novel findings and redefinition of the critically deleted regions. Am J Med Genet Part A 2012;158a:2266–71.
Vissers LE, Cox TC, Maga AM, Short KM, Wiradjaja F, Janssen IM, et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet 2011;7:e1002278.
Recalcati MP, Bellini M, Norsa L, Ballarati L, Caselli R, Russo S, et al. Complex rearrangement involving 9p deletion and duplication in a syndromic patient: genotype/phenotype correlation and review of the literature. Gene 2012;502:40–5.
Kawara H, Yamamoto T, Harada N, Yoshiura K, Niikawa N, Nishimura A, et al. Narrowing candidate region for monosomy 9p syndrome to a 4.7-Mb segment at 9p22.2-p23. Am J Med Genet Part A 2006;140:373–77.
Sousa SB, Hennekam RC. Phenotype and genotype in Nicolaides-Baraitser syndrome. Am J Med Genet Part C, Semin Med Genet. 2014;166c:302–14.
Willemsen MH, Vulto-van Silfhout AT, Nillesen WM, Wissink-Lindhout WM, van Bokhoven H, Philip N, et al. Update on Kleefstra syndrome. Mol Syndromol 2012;2:202–12.
Iacono G, Dubos A, Meziane H, Benevento M, Habibi E, Mandoli A, et al. Increased H3K9 methylation and impaired expression of Protocadherins are associated with the cognitive dysfunctions of the Kleefstra syndrome. Nucleic Acids Res 2018;46:4950–65.
Ribeiro AL, Silva RD, Foyn H, Tiago MN, Rathore OS, Arnesen T, et al. Naa50/San-dependent N-terminal acetylation of Scc1 is potentially important for sister chromatid cohesion. Sci Rep 2016;6:39118.
Oliveira J, Dias C, Redeker E, Costa E, Silva J, Reis Lima M, et al. Development of NIPBL locus-specific database using LOVD: from novel mutations to further genotype-phenotype correlations in Cornelia de Lange Syndrome. Hum Mutat 2010;31:1216–22.
Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 2007;80:485–94.
Acknowledgements
We would like to thank the patients and their families, and especially the Brazilian Association of Cornelia de Lange Syndrome (CdLS Brazil) for participating in this study. We also thank N. Watanabe, T. Miyama, M. Sato, S. Sugimoto, and K. Takabe for their technical assistance. This work was supported by Japan Agency for Medical Research and Development (AMED) under grant numbers JP18ek0109280, JP18dm0107090, JP18ek0109301, JP18ek0109348, and JP18kk0205001; by Japan Society for the Promotion of Science (JSPS) KAKENHI under grant numbers JP17H01539, JP16H05357, JP16H06254, JP17K16132, JP17K10080, JP17K15630, and JP17H06994; by the Ministry of Health, Labour, and Welfare; and by the Takeda Science Foundation. We thank Sarah Williams, PhD, from Edanz Group (www.edanzediting.com) for editing a draft of this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Aoi, H., Mizuguchi, T., Ceroni, J. et al. Comprehensive genetic analysis of 57 families with clinically suspected Cornelia de Lange syndrome. J Hum Genet 64, 967–978 (2019). https://doi.org/10.1038/s10038-019-0643-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0643-z