Abstract
Proper diagnosis of mild inflicted traumatic brain injury (ITBI) is difficult; children often present without a history of trauma and with nonspecific symptoms, such as vomiting. Previous studies suggest that biomarkers may be able to screen for brain injury in this population, but these studies focused on only a few biomarkers. We hypothesized that using multiplex bead technology we would be able to identify multiple differences in the serum biomarker profile between in children with ITBI and those without brain injury. We compared the concentrations of 44 serum biomarkers in 16 infants with mild ITBI and 20 infants without brain injury. There were significant group differences in the concentrations of nine of the 44 markers. Vascular cellular adhesion molecule (VCAM) (p < 0.00) and IL-6 (IL-6) (p < 0.00) had the most significant group differences; IL-6 was higher after ITBI, whereas VCAM was lower. Using VCAM and IL-6 in classification algorithms, we could discriminate the groups with a sensitivity and specificity of 87% and 90%, respectively. The results suggest significant changes in the serum biomarker profile after mild ITBI. Future research is needed to determine whether these biomarkers can screen for brain injury in infants with nonspecific symptoms.
Similar content being viewed by others
Main
Inflicted traumatic brain injury (ITBI) is a leading cause of death from traumatic brain injury (TBI) in infants (2,3). Proper diagnosis of ITBI is often difficult even for experienced, astute physicians because caretakers rarely provide a history of trauma (4,5), children present with nonspecific symptoms, such as vomiting or fussiness (6) and physical examination can be normal (7,8). As a result, misdiagnosis is common and can have catastrophic medical consequences (9,10). The importance of timely diagnosis of ITBI cannot be overemphasized: if not recognized, child abuse is an escalating form of trauma that often ends in death or disability (11).
The frequency with which ITBI is misdiagnosed and the resulting morbidity and mortality are compounded by the lack of a well-established screening test to help physicians identify children who might benefit from evaluation with cranial computed tomography (CT). We previously reported on the possible use of serum biomarkers as screening tools for infants at increased risk of missed ITBI—infants without a history of trauma and with nonspecific symptoms (12–15). We focused on three well-established and relatively brain-specific markers: neuron-specific enolase, S100B, and myelin-basic protein. Our study evaluating these markers as screening tools for ITBI in high-risk infants showed that a combination of neuron-specific enolase and myelin-basic protein was 79% sensitive and 70% specific for brain injury (14). Contrary to previous literature in adults and older children, S100B was not specific for brain injury in infants.
Biomarker concentrations in our previous studies have been measured using ELISA. ELISA allows for measurement of only one biomarker at a time and requires up to 100 μL of serum for each biomarker. In young children, it is difficult to collect large sample volumes and 100 μL may be the entire sample.
Multiplex bead technology employs uniquely labeled microspheres processed as an immunoassay tagged with fluorescent labeled markers. A flow cytometer equipped with a complex optic system identifies and quantifies each protein. Advantages of this technique are the small volume required—<100 μL can provide results for up to 100 proteins—and the fact that because it measures multiple markers simultaneously, it is less labor-intensive than ELISA. A recent study in our laboratory was the first to use this technology in cerebrospinal fluid (CSF) from children with severe TBI (16). To our knowledge, it has not been used previously with serum from pediatric TBI patients. Unlike our previous studies, which have focused on brain-specific markers, the proteins measured using multiplex beads are markers of systemic response to tissue injury.
Therefore, the objectives of this study were to explore the feasibility of using multiplex bead assays to measure serum biomarkers in children with ITBI and compare multiple biomarkers in children with mild ITBI (cases) to those without brain injury (controls). We also sought to assess the potential screening value of nonbrain specific biomarkers. We did not seek to evaluate the use of these markers to determine the etiology (e.g. abuse versus not abuse) of TBI, but only to characterize differences in the biomarker profile between children with and without brain injury. We hypothesized that there would be multiple differences in the serum biomarker profile between these groups.
METHODS
Subjects.
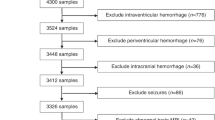
The study was exempt by the University of Pittsburgh Institutional Review Board because it used blood collected with consent as part of previously approved protocols. Children were eligible as cases if they presented with a Glasgow Coma Scale score of 15 and were given a diagnosis of ITBI by the Children's Hospital of Pittsburgh Child Protection Team. Defining whether an injury is inflicted based on the conclusion of a Child Protection Team is frequently used in studies of ITBI (2,9,17). When trauma was not initially recognized, as was true in most cases, the Glasgow Coma Scale score was assigned retrospectively based on emergency department documentation. In each ITBI case, the time of injury was unknown and was defined as the time at which the child began exhibiting symptoms. If that was unknown, then patient presented to the emergency department for care was used as the time of injury. CT scans were evaluated by a pediatric neuroradiologist as part of routine clinical care. There were two groups of controls. For both groups, children were <1 y of age, afebrile (defined as temperature <38.3°C), had no diarrhea and no history of trauma. The first group presented to Children's Hospital of Pittsburgh emergency department with symptoms, which would place them at increased risk for a brain injury as previously defined by several studies evaluating infants at risk for missed ITBI (9,14). These symptoms included vomiting, fussiness, an apparent-life threatening event, or a possible seizure. As part of their evaluation, these infants had blood drawn and a head CT that was normal. The second group went to the operating room for elective outpatient surgery, which required general anesthesia (e.g. hernia repair) and had no history of any neurologic concerns. In these cases, blood was collected at the time vascular access was obtained. Enrollment was not consecutive and was based on availability of the investigators. Some infants in the second control group did not have a head CT because it was not clinically indicated.
Biomarker measurement.
Five separate multiplex bead array assays (BioSource 38-plex, Linco CVD1 4-plex, Linco CVD2 1-plex for haptoglobin, Linco CVD2 1-plex for fibrinogen, and a bead developed at the University of Pittsburgh Cancer Institute to measure heat shock protein 70) were used to measure 44 markers according to the manufacturers' instructions. The markers measured included heat shock protein 70, matrix metallopeptidase-9 (MMP-9), plasminogen activator inhibitor-1, intracellular adhesion molecule (ICAM), vascular cellular adhesion molecule (VCAM), fibrinogen, haptoglobin, and 38 cytokines [IL-1α and β, IL-2, 4, 5, 6, 7, 8, 10, 12, 13, 15, and 17, and the associated receptors for IL1, and 2, tumor necrosis factor (TNF α), and TNF receptor-1 (TNFR1) and 2 (TNFR2), interferon-α (IFN-α) and IFNγ, granulocyte-macrophage colony-stimulating factor, granulocyte- colony-stimulating factor, macrophage inflammatory protein 1α and 1β, IFN inducing protein-10, monokine induced by γ-interferon, eotaxin, RANTES, monocyte chemoattractant protein1, 2, and 3, death receptor 5, epidermal growth factor 1, fibroblast growth factor β, hepatocyte growth factor (HGF), VEGF, and growth-regulated oncogene α]. Marker concentrations were quantified when they were within the appropriate limits of each standard curve.
Statistical analysis.
Descriptive statistics and graphical displays (i.e. dot plots) were used to compare the demographics of the groups and the distribution of each marker in each group. Spearman's (nonparametric) correlation was calculated to quantify the relationships between each pair of markers. The Wilcoxon rank-sum test was used to evaluate the significance of differences in marker expression between groups. Semipartial correlations from a logistic regression model were used to evaluate the unique contribution of each variable to the prediction of group. All p values are two-sided, and <0.05 was considered statistically significant. These statistical tests were conducted using SPSS Software (Version 14, SPSS Inc, Chicago, IL).
Three different algorithms—linear classification (18–20), five nearest neighbors (21) and tree classification (22)—were used to separate subjects by group. The linear classification algorithm calculates a line so that most of one group lies on one side, whereas most of the other group lies on the other side. An unknown patient is classified based on which the side of the line it lies. The five nearest neighbor's algorithm classifies the group of an unknown patient by finding the five patients in a master dataset, which are nearest to the unknown patient and selecting the group, that most of these patients belong to. The tree classifier algorithm builds a binary tree of decision nodes and associates each terminal node with either the group of cases or controls. These three algorithms were performed using Matlab (Version 7.0.0, Mathworks Inc.).
RESULTS
Patient demographics.
The concentrations of 44 biomarkers were compared in 16 cases and 20 controls. Of the 20 controls, 15 (75%) presented with nonspecific symptoms as described above and 5 (25%) presented for an out-patient surgical procedure; these groups are analyzed as a single control group because, as discussed below, there was no difference in any of the biomarker concentrations between the groups. There were no differences in the proportion of male or white patients between cases and controls. The mean (SD) age of the cases was slightly greater than that of controls [7.5 (7.3) versus 3.4 (2.6) mo, p = 0.05]. The mean (SD) time after injury at which the blood sample was collected for cases was 26.0 (25.2) h. All cases had a head CT as part of clinical care: 12 of 16 (75%) had an acute subdural hemorrhage(s), 3 (19%) also had probable or definite chronic subdural hemorrhage(s), 6 (38%) had an intraparenchymal contusion or hemorrhage, 3 (19%) had a skull fracture, and 4 (25%) had evidence of early cerebral edema. Of the 16 cases, 38% (6/16) had extra-cranial injuries, all of which were rib and/or metaphyseal fractures.
Serum biomarker concentrations.
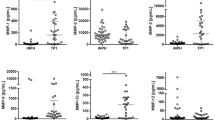
Of the 44 markers measured, 21 markers had concentrations which were undetectable in >50% of subjects with no group differences, 14 markers were detectable in both cases and controls, but without group differences and nine markers had significant differences between cases and controls (Table 1). There were no differences in the concentrations of any of the markers between the two control groups, and thus they were merged into a single control group. The markers with significant differences between cases and controls were VCAM (p < 0.00), IL-12 (p = 0.02), MMP-9 (p < 0.00), ICAM (p = 0.03), eotaxin (p = 0.04), HGF (p < 0.00), TNFR2 (p = 0.05), IL-6 (p < 0.00), and fibrinogen (p = 0.00) (Fig. 1). Markers that were higher in cases were MMP 9, HGF, fibrinogen, and IL-6. Markers that had higher concentration in controls were ICAM, VCAM, IL-12, eotaxin, and TNFR2. There was no difference in biomarker concentrations between cases with and without extra-cranial injuries.
Because of concern of the effect of multiple comparisons on the type I error rate, we calculated semipartial correlations from a single logistic model using each of the 23 markers. The biomarkers with significant partial correlations were the same as those with differences by Wilcoxon's rank sum with one exception; TNFR2 had a p = 0.05 by Wilcoxon's rank sum, but a nonsignificant partial correlation. The biomarkers with significant partial correlations were then used in a regression analysis. This produces a single omnibus χ2 test, which fixes the overall type 1 error level at 0.05. Using these markers, the χ2 was significant (p < 0.00), the model fit was acceptable (Cox and Snell R2 = 0.575) and the correct classification rate was 88%. These data therefore support our original conclusions and suggest that with the possible exception of TNFR2, the markers identified previously are good candidates to be evaluated in a prospective validation.
Correlation between biomarkers.
Analysis of the correlations between each of the nine markers with significant differences between cases and controls showed multiple significant correlations (Table 2). The strongest positive correlations were between ICAM and VCAM (r = 0.49, p < 0.00), MMP-9 and HGF (r = 0.86, p < 0.00), ICAM and TNFR2 (r = 0.57, p < 0.00), and IL-6 and eotaxin (r = 0.76, p = 0.01). The strongest inverse correlation was between MMP-9 and VCAM (r = −0.45, p < 0.01).
Classification algorithms.
Using VCAM and IL-6—the two markers with the most significant differences between groups in Wilcoxon's rank sum test—cases and controls were separated using the algorithms described above. Only two markers were evaluated to avoid over-fitting with this small sample. The classification (with leave-one-out cross-validation) using only VCAM and IL-6 shows a sensitivity of 87% and specificity of 90% for discrimination of cases and controls (Fig. 2). The sensitivity and specificity of all three algorithms were virtually identical, supporting the robustness of the results.
A scatter plot of the normalized concentrations of IL-6 and vascular cellular adhesion molecule (VCAM). Scatter plot concentrations were normalized to allow for simultaneous visualization of two biomarkers with concentrations, which were of different orders of magnitude. The median concentration for each marker was set to zero and other values were calculated as the number of SD above or below the median. Two extreme IL-6 outliers—one in each group—were removed for purposes of the scatter plot. bull; = controls, ○ = cases.
Although the sensitivity and specificity of the combination of VCAM and IL-6 was high, we repeated the five nearest neighbor and separating hyperplane classification algorithms using IL-6 and MMP-9, the two markers that were most significantly increased after TBI. The reason for repeating the analysis with MMP-9 and IL-6 was twofold; first, the decrease in VCAM concentrations after TBI was unexpected on a physiologic basis and thus we questioned whether it would be clinically acceptable as a marker of injury and second, on a clinical basis, it is more difficult to use a biomarker whose concentration decreases, rather than increases, after injury. This is particularly true when the marker's concentration is in the nanogram per milliliter range. The classification using the five nearest neighbor algorithm with MMP-9 and IL-6 shows a sensitivity of 81% and specificity of 90%. The separating hyperplane algorithm had the same specificity with a sensitivity of 94%.
DISCUSSION
This is the first study to use multiplex bead technology to simultaneously measure serum biomarkers after pediatric TBI of any etiology. Our ability to measure 44 markers allowed for identification of multiple novel biomarkers that are either increased or decreased after TBI compared with controls and suggests that this technique is an efficient method to identify new biomarkers of interest. This study was also unique because the cases were children with mild TBI; most of the TBI biomarker literature focuses on patients with moderate to severe TBI.
The significant group differences for nine of the 44 biomarkers tested demonstrates that even children with mild ITBI have a biomarker profile, which is distinct from that of similarly aged children without TBI. The differences in the biomarker profile suggest that these markers may be able to assist in the biochemical discrimination of two groups of children, which can be difficult to distinguish clinically: young children with nonspecific symptoms because of common childhood illnesses and those with the same symptoms because of TBI. It is important that our findings and particularly the derived classification algorithms be prospectively validated in a different cohort of patients.
It is important to recognize that we did not evaluate the use of these markers to discriminate etiology (e.g. abuse versus not abuse) of TBI, but only to identify the presence of TBI. Thus, use of these markers would not change the way in which ITBI is diagnosed, but would only be used to alert the treating physician to the possible presence of brain injury and thereby prompt neuroimaging in a patient who would otherwise be discharged without it. The similar patterns for many biomarkers in children with TBI of different etiologies is supported by a recent study by Buttram et al, (16) which demonstrated no differences in the CSF cytokine response between children with ITBI and those with noninflicted TBI (16). Similarly, because the markers evaluated in this study are not brain-specific, they would be unlikely to distinguish patients with TBI from those with noncranial trauma. Hensler et al (23) measured serum concentrations of several biomarkers including IL-6 and 10 in adults with severe TBI and compared them to concentrations in adults with multiple significant noncranial injuries, and found no difference in the concentrations between groups. In our population of interest—well-appearing infants at increased risk of abuse—severe extra-cranial injuries are unlikely. This is supported by our own data; none of the extra-cranial injuries in the cases was severe and there was no difference in the biomarker concentrations between cases with and without extra-cranial injuries.
Although several of these markers have been shown to be increased in the presence of fever (24,25) and in a wide variety of pediatric illnesses, including enteroviral meningitis (IL-6) (26), influenza (IL-6) (25), and rotatvirus (IL-6, IL-10, and IFN-gamma) (27), we do not believe this a clinical limitation given our target population. The population at risk for ITBI that would be targeted by a biomarker screen is afebrile infants with nonspecific symptoms.
We were unable to assess whether the increase in serum concentrations of the biomarkers was due to increased leakage from brain into blood because of blood-brain barrier dysfunction or the result of a systemic response to brain injury. Kossman et al (28) performed a study in which 20 adults with severe TBI had simultaneous CSF and serum IL-6 measurements as well as assessment of blood-brain barrier dysfunction. Their results suggest that although CSF is the origin of serum IL-6 on day 1 after injury, the source of IL-6 at later time points is extra-cerebral and released as part of the systemic response to injury. This has not been addressed in pediatric TBI.
Our finding that ICAM, VCAM IL-12, and TNFR2 concentrations were lower in children with ITBI compared with controls is contrary to previous CSF literature, which showed increased concentrations of these markers after TBI (29–33). There are no published studies of serum IL-12 concentrations or CSF or serum eotaxin concentrations after human TBI. The limited literature related to serum ICAM and TNFR2 concentrations after human TBI includes only adult patients with severe TBI; two studies demonstrated increased serum ICAM (31) and TNFR2 (34) concentrations and a third (35) demonstrated no difference in initial serum ICAM concentrations, but significantly higher concentrations 96 h after TBI. It will be important to analyze serum concentrations of these markers in children with TBI of different severities and etiologies to better assess whether these differences are related to severity or etiology of injury (e.g. abuse versus not abuse) or patient age (e.g. adult versus pediatric). Many factors such as the stress response could initiate a systemic injury response in the setting of ITBI. It will also be important to assess the possible contribution of timing to these biomarker differences. It is not surprising that the mean time of sample collection for the ITBI cases was 26 h after injury/onset of symptoms, because there is often a delay in seeking medical care in cases of abuse. The effect of this delay on the biomarker concentrations will need to be evaluated using serial samples collected in infants and young children with noninflicted TBI in whom early samples are available.
The increased serum MMP-9, IL-6, and fibrinogen concentrations are consistent with previous TBI literature in adults (28,36,37). There is one report showing increased CSF HGF concentrations after pediatric TBI. In this report, increased HGF concentrations were associated with young age and poor outcome (Fink, E, et al. CSF analysis of HGF concentrations in infants and children after TBI, Society of Critical Care Medicine 31st Critical Care Congress. January 26-30, 2002, San Diego, California, abstract: A140).
The study's most significant limitation was the modest sample size. We were unable to simultaneously evaluate more than two markers in the classification tree. Although the number of cases is small from a statistical standpoint, it is relatively large from a clinical perspective; in a large children's hospital with a level I trauma center and a large child abuse program, it took several years to enroll 16 children with ITBI and a Glasgow Coma Scale score of 15. This highlights both the difficulty obtaining consent from this population and the difficulty of recognizing mild ITBI. Epidemiologically, the majority of infants diagnosed with ITBI have severe ITBI; this is unlikely to be the result of a marked difference in the epidemiology of ITBI and noninflicted TBI, but rather the result of the clinical problem we are trying to address, the difficulty in recognizing more mild forms of ITBI. Despite this limitation, we were able to classify subjects with >85% accuracy using VCAM and IL-6.
Several of the control patients did not have a head CT as part of clinical care; as a result, we cannot rule out the possibility that one or more of them had brain injury. If, however, any of the controls had an undetected TBI, this would make the difference between the groups artificially smaller. We believe it is unethical to perform a head CT in a child in whom there is no clinical indication (e.g. for research purposes) because of the radiation risk associated with head CT in young children (38–40).
In summary, this is the first study to multiplex bead technology to simultaneously assess multiple serum biomarkers in children with ITBI. We have identified several novel biomarkers, which are either increased or decreased after mild pediatric ITBI compared with controls. The results support our hypothesis that there are differences in the serum concentrations of nonbrain specific biomarkers in children with ITBI compared with those without brain injury. A panel of biomarker, which includes a combination of brain-specific and tissue injury response markers, may be better able to discriminate patients with and without brain injury than a panel of either type of marker alone. A prospective validation using a larger patient population is essential to allow for potentially exciting classification analyses using a greater number of both brain-specific and nonbrain specific biomarkers.
Abbreviations
- CSF:
-
cerebrospinal fluid
- CT:
-
computed tomography
- HGF:
-
hepatocyte growth factor
- ITBI:
-
inflicted traumatic brain injury
- MMP-9:
-
matrix metallopeptidase-9
- TBI:
-
traumatic brain injury
- VCAM:
-
vascular cellular adhesion molecule
References
Deleted in proof
Keenan HT, Runyan DK, Marshall SW, Nocera MA, Merten DF, Sinal SH 2003 A population-based study of inflicted traumatic brain injury in young children. JAMA 290: 621–626
Duhaime AC, Christian CW, Rorke LB, Zimmerman RA 1998 Nonaccidental head injury in infants-the “shaken-baby syndrome”. N Engl J Med 338: 1822–1829
O'Neill JA Jr, Meacham WF, Griffin JP, Sawyers JL 1973 Patterns of injury in the battered child syndrome. J Trauma 13: 332–339
Ettaro L, Berger RP, Songer T 2004 Abusive head trauma in young children: characteristics and medical charges in a hospitalized population. Child Abuse Negl 28: 1099–1111
Duhaime AC, Partington MD 2002 Overview and clinical presentation of inflicted head injury in infants. Neurosurg Clin N Am 13: 149–154 v
Haviland J, Russell RI 1997 Outcome after severe non-accidental head injury. Arch Dis Child 77: 504–507
Morris MW, Smith S, Cressman J, Ancheta J 2000 Evaluation of infants with subdural hematoma who lack external evidence of abuse. Pediatrics 105: 549–553
Jenny C, Hymel KP, Ritzen A, Reinert SE, Hay TC 1999 Analysis of missed cases of abusive head trauma. JAMA 281: 621–626
Ewing-Cobbs L, Kramer L, Prasad M, Canales DN, Louis PT, Fletcher JM, Vollero H, Landry SH, Cheung K 1998 Neuroimaging, physical, and developmental findings after inflicted and noninflicted traumatic brain injury in young children. Pediatrics 102: 300–307
Ellaway BA, Payne EH, Rolfe K, Dunstan FD, Kemp AM, Butler I, Sibert JR 2004 Are abused babies protected from further abuse?. Arch Dis Child 89: 845–846
Berger RP, Kochanek PM, Pierce MC 2004 Biochemical markers of brain injury: could they be used as diagnostic adjuncts in cases of inflicted traumatic brain injury?. Child Abuse Negl 28: 739–754
Berger RP, Adelson PD, Pierce MC, Dulani T, Cassidy LD, Kochanek PM 2005 Serum neuron-specific enolase, S100B, and myelin basic protein concentrations after inflicted and noninflicted traumatic brain injury in children. J Neurosurg 103: 61–68
Berger RP, Dulani T, Adelson PD, Leventhal JM, Richichi R, Kochanek PM 2006 Identification of inflicted traumatic brain injury in well-appearing infants using serum and cerebrospinal markers: a possible screening tool. Pediatrics 117: 325–332
Berger RP, Pierce MC, Wisniewski SR, Adelson PD, Clark RS, Ruppel RA, Kochanek PM 2002 Neuron-specific enolase and S100B in cerebrospinal fluid after severe traumatic brain injury in infants and children. Pediatrics 109: E31
Buttram SD, Wisniewski SR, Jackson EK, Adelson PD, Feldman K, Bayir H, Berger RP, Clark RS, Kochanek PM 2007 Multiplex assessment of cytokine and chemokine levels in cerebrospinal fluid following severe pediatric traumatic brain injury: effects of moderate hypothermia. J Neurotrauma 24: 1707–1718
Hymel KP, Abshire TC, Luckey DW, Jenny C 1997 Coagulopathy in pediatric abusive head trauma. Pediatrics 99: 371–375
Mangasarian OL, Street WN, Wolberg WH 1995 Breast cancer diagnosis and prognosis via linear programming. Oper Res 43: 570–577
Mangasarian O, Setiono R, Wolberg W 1990 Pattern recognition via linear programming: theory and applications to medical diagnosis. In: Coleman F, Li Y (eds) Large Scale Numerical Optimization.. Society for Industrial and Applied Mathematics (SIAM) Publications, University of Michigan, Ann Arbor, pp 22–30
Mangasarian O 1965 Linear and non-linear separation of patterns by linear programming. Oper Res 13: 444–452
Friedman J, Hastie T, Tibshirani R 2001 The Elements of Statistical Learning, Data Mining Inference and Prediction. New York: Springer, pp 415–420
Breiman J, Friedman R, Stone C 1984 Classification and Regression Trees. New York: Chapman & Hall, pp 18–58
Hensler T, Sauerland S, Bouillon B, Raum M, Rixen D, Helling HJ, Andermahr J, Neugebauer EA 2002 Association between injury pattern of patients with multiple injuries and circulating levels of soluble tumor necrosis factor receptors, interleukin-6 and interleukin-10, and polymorphonuclear neutrophil elastase. J Trauma 52: 962–970
Engel A, Kern WV, Murdter G, Kern P 1994 Kinetics and correlation with body temperature of circulating interleukin-6, interleukin-8, tumor necrosis factor alpha and interleukin-1 beta in patients with fever and neutropenia. Infection 22: 160–164
Kaiser L, Fritz RS, Straus SE, Gubareva L, Hayden FG 2001 Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J Med Virol 64: 262–268
Sato M, Hosoya M, Honzumi K, Watanabe M, Ninomiya N, Shigeta S, Suzuki H 2003 Cytokine and cellular inflammatory sequence in enteroviral meningitis. Pediatrics 112: 1103–1107
Jiang B, Snipes-Magaldi L, Dennehy P, Keyserling H, Holman RC, Bresee J, Gentsch J, Glass RI 2003 Cytokines as mediators for or effectors against rotavirus disease in children. Clin Diagn Lab Immunol 10: 995–1001
Kossmann T, Hans VH, Imhof HG, Stocker R, Grob P, Trentz O, Morganti-Kossmann C 1995 Intrathecal and serum interleukin-6 and the acute-phase response in patients with severe traumatic brain injuries. Shock 4: 311–317
Whalen MJ, Carlos TM, Kochanek PM, Wisniewski SR, Bell MJ, Carcillo JA, Clark RS, DeKosky ST, Adelson PD 1998 Soluble adhesion molecules in CSF are increased in children with severe head injury. J Neurotrauma 15: 777–787
Pleines UE, Morganti-Kossmann MC, Rancan M, Joller H, Trentz O, Kossmann T 2001 S-100 beta reflects the extent of injury and outcome, whereas neuronal specific enolase is a better indicator of neuroinflammation in patients with severe traumatic brain injury. J Neurotrauma 18: 491–498
Pleines UE, Stover JF, Kossmann T, Trentz O, Morganti-Kossmann MC 1998 Soluble ICAM-1 in CSF coincides with the extent of cerebral damage in patients with severe traumatic brain injury. J Neurotrauma 15: 399–409
Stahel PF, Kossmann T, Joller H, Trentz O, Morganti-Kossmann MC 1998 Increased interleukin-12 levels in human cerebrospinal fluid following severe head trauma. Neurosci Lett 249: 123–126
Amick JE, Yandora KA, Bell MJ, Wisniewski SR, Adelson PD, Carcillo JA, Janesko KL, DeKosky ST, Carlos TM, Clark RS, Kochanek PM 2001 The Th1 versus Th2 cytokine profile in cerebrospinal fluid after severe traumatic brain injury in infants and children. Pediatr Crit Care Med 2: 260–264
Goodman JC, Robertson CS, Grossman RG, Narayan RK 1990 Elevation of tumor necrosis factor in head injury. J Neuroimmunol 30: 213–217
McKeating EG, Andrews PJ, Mascia L 1998 The relationship of soluble adhesion molecule concentrations in systemic and jugular venous serum to injury severity and outcome after traumatic brain injury. Anesth Analg 86: 759–765
Suehiro E, Fujisawa H, Akimura T, Ishihara H, Kajiwara K, Kato S, Fujii M, Yamashita S, Maekawa T, Suzuki M 2004 Increased matrix metalloproteinase-9 in blood in association with activation of interleukin-6 after traumatic brain injury: influence of hypothermic therapy. J Neurotrauma 21: 1706–1711
Conti A, Sanchez-Ruiz Y, Bachi A, Beretta L, Grandi E, Beltramo M, Alessio M 2004 Proteome study of human cerebrospinal fluid following traumatic brain injury indicates fibrin(ogen) degradation products as trauma-associated markers. J Neurotrauma 21: 854–863
Browning JG, Reed MJ, Wilkinson AG, Beattie T 2005 Imaging infants with head injury: effect of a change in policy. Emerg Med J 22: 33–36
Brenner D, Elliston C, Hall E, Berdon W 2001 Estimated risks of radiation-induced fatal cancer from pediatric CT. AJR Am J Roentgenol 176: 289–296
Brody AS, Frush DP, Huda W, Brent RL 2007 Radiation risk to children from computed tomography. Pediatrics 120: 677–682
Acknowledgements
We thank Adele Marrangoni for technical expertise and Marci Provins, Ranjodh Singh, and Diane Weidner for administrative assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by University of Pittsburgh Center for Injury Research and Control [R.B., P.K.].
Rights and permissions
About this article
Cite this article
Berger, R., Ta'asan, S., Rand, A. et al. Multiplex Assessment of Serum Biomarker Concentrations in Well-Appearing Children With Inflicted Traumatic Brain Injury. Pediatr Res 65, 97–102 (2009). https://doi.org/10.1203/PDR.0b013e31818c7e27
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e31818c7e27
This article is cited by
-
Mild abusive head injury: diagnosis and pitfalls
Child's Nervous System (2022)
-
Traumatic brain injury biomarkers in pediatric patients: a systematic review
Neurosurgical Review (2022)
-
Fluid proteomics of CSF and serum reveal important neuroinflammatory proteins in blood–brain barrier disruption and outcome prediction following severe traumatic brain injury: a prospective, observational study
Critical Care (2021)
-
Inflammation in epileptogenesis after traumatic brain injury
Journal of Neuroinflammation (2017)
-
Fluid biomarkers for mild traumatic brain injury and related conditions
Nature Reviews Neurology (2016)