Abstract
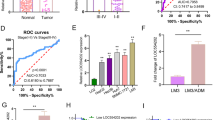
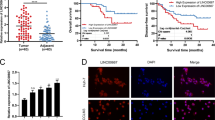
It has long been known that males are more susceptible than females to hepatocellular carcinoma (HCC), but the reason remains elusive. In this study, we investigated the expression and function of the long noncoding RNA FTX (lnc-FTX), an X-inactive-specific transcript (XIST) regulator transcribed from the X chromosome inactivation center, in both HCC and HCC gender disparity. lnc-FTX is expressed at higher levels in female livers than in male livers and is significantly downregulated in HCC tissues compared with normal liver tissues. Patients with higher lnc-FTX expression exhibited longer survival, suggesting that lnc-FTX is a useful prognostic factor for HCC patients. lnc-FTX inhibits HCC cell growth and metastasis both in vitro and in vivo. Mechanistically, lnc-FTX represses Wnt/β-catenin signaling activity by competitively sponging miR-374a and inhibits HCC cell epithelial–mesenchymal transition and invasion. In addition, lnc-FTX binds to the DNA replication licensing factor MCM2, thereby impeding DNA replication and inhibiting proliferation in HCC cells. In conclusion, these findings suggest that lnc-FTX may act as a tumor suppressor in HCC through physically binding miR-374a and MCM2. It may also be one of the reasons for HCC gender disparity and may potentially contribute to HCC treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Abbreviations
- AFP:
-
α-fetoprotein
- EdU:
-
5-ethynyl-2′-deoxyuridine
- EMT:
-
epithelial–mesenchymal transition
- HCC:
-
hepatocellular carcinoma
- lnc-FTX:
-
long noncoding RNA FTX
- lncRNA:
-
long non-coding RNA
- MCM2:
-
minichromosome maintenance complex component 2
- XCI:
-
X chromosome inactivation
- XIST:
-
X-inactive-specific transcript
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A . Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108.
El-Serag HB, Rudolph KL . Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132: 2557–2576.
Yang D, Hanna DL, Usher J, LoCoco J, Chaudhari P, Lenz HJ et al. Impact of sex on the survival of patients with hepatocellular carcinoma: a surveillance, epidemiology, and end results analysis. Cancer 2014; 120: 3707–3716.
Buch SC, Kondragunta V, Branch RA, Carr BI . Gender-based outcomes differences in unresectable hepatocellular carcinoma. Hepatol Int 2008; 2: 95–101.
Dohmen K, Shigematsu H, Irie K, Ishibashi H . Longer survival in female than male with hepatocellular carcinoma. J Gastroenterol Hepatol 2003; 18: 267–272.
Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007; 317: 121–124.
Shimizu I, Kohno N, Tamaki K, Shono M, Huang HW, He JH et al. Female hepatology: favorable role of estrogen in chronic liver disease with hepatitis B virus infection. World J Gastroenterol 2007; 13: 4295–4305.
Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng IO et al. Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell 2014; 25: 49–63.
Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L et al. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res 2002; 12: 894–908.
Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet 2011; 20: 705–718.
Reiche K, Kasack K, Schreiber S, Luders T, Due EU, Naume B et al. Long non-coding RNAs differentially expressed between normal versus primary breast tumor tissues disclose converse changes to breast cancer-related protein-coding genes. PLoS One 2014; 9: e106076.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP . A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011; 147: 344–357.
Chi SW, Zang JB, Mele A, Darnell RB . Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009; 460: 479–486.
Zinchuk V, Zinchuk O, Okada T . Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem 2007; 40: 101–111.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142: 409–419.
Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF . Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009; 139: 719–730.
Shechter D, Ying CY, Gautier J . DNA unwinding is an Mcm complex-dependent and ATP hydrolysis-dependent process. J Biol Chem 2004; 279: 45586–45593.
Giaginis C, Vgenopoulou S, Vielh P, Theocharis S . MCM proteins as diagnostic and prognostic tumor markers in the clinical setting. Histol Histopathol 2010; 25: 351–370.
Reena RM, Mastura M, Siti-Aishah MA, Munirah MA, Norlia A, Naqiyah I et al. Minichromosome maintenance protein 2 is a reliable proliferative marker in breast carcinoma. Ann Diagn Pathol 2008; 12: 340–343.
Giaginis C, Georgiadou M, Dimakopoulou K, Tsourouflis G, Gatzidou E, Kouraklis G et al. Clinical significance of MCM-2 and MCM-5 expression in colon cancer: association with clinicopathological parameters and tumor proliferative capacity. Dig Dis Sci 2009; 54: 282–291.
Sun M, Wu G, Li Y, Fu X, Huang Y, Tang R et al. Expression profile reveals novel prognostic biomarkers in hepatocellular carcinoma. Front Biosci (Elite Ed) 2010; 2: 829–840.
Quaglia A, McStay M, Stoeber K, Loddo M, Caplin M, Fanshawe T et al. Novel markers of cell kinetics to evaluate progression from cirrhosis to hepatocellular carcinoma. Liver Int 2006; 26: 424–432.
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest 2013; 123: 566–579.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
De Craene B, Berx G . Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110.
Batista PJ, Chang HY . Long noncoding RNAs: cellular address codes in development and disease. Cell 2013; 152: 1298–1307.
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut 2015.
Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology 2013; 58: 739–751.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–681.
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015; 16: 413–425.
Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG et al. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008; 22: 3172–3183.
Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y et al. Structure and activity of putative intronic miRNA promoters. RNA 2010; 16: 495–505.
Siomi H, Siomi MC . Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 2010; 38: 323–332.
Hartwell HJ, Petrosky KY, Fox JG, Horseman ND, Rogers AB . Prolactin prevents hepatocellular carcinoma by restricting innate immune activation of c-Myc in mice. Proc Natl Acad Sci USA 2014; 111: 11455–11460.
Shi L, Feng Y, Lin H, Ma R, Cai X . Role of estrogen in hepatocellular carcinoma: is inflammation the key? J Transl Med 2014; 12: 93.
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010; 40: 939–953.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344: 310–313.
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015; 18: 603–610.
Acknowledgements
We thank Dr Le Qu and Mr Zhi-peng Xu from Department of Urology, Shanghai Changzheng Hospital and Dr Sheng-Xian Yuan and Dr Qi-Fei Tao from the Third Department of Hepatic Surgery, Eastern Hepatobiliary Hospital for technical assistance. This work was supported by grants from the State Key Program of National Natural Science Foundation of China (grant nos 81330037), the National Key Basic Research Program of China (973 Program) (grant nos 2015CB554004), the National Natural Science Foundation of China (grant nos 31201025, 81301831, 81301692, 81472691 and 81402269, and the National Natural Science Foundation of Shanghai (13ZR1448300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, F., Yuan, JH., Huang, JF. et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 35, 5422–5434 (2016). https://doi.org/10.1038/onc.2016.80
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.80
This article is cited by
-
Connecting the mechanisms of tumor sex differences with cancer therapy
Molecular and Cellular Biochemistry (2024)
-
The pluripotent factor OCT4A enhances the self-renewal of human dental pulp stem cells by targeting lncRNA FTX in an LPS-induced inflammatory microenvironment
Stem Cell Research & Therapy (2023)
-
Emerging roles of long non-coding RNA FTX in human disorders
Clinical and Translational Oncology (2023)
-
Long noncoding RNA Ftx regulates the protein expression profile in HCT116 human colon cancer cells
Proteome Science (2022)
-
MCM2 in human cancer: functions, mechanisms, and clinical significance
Molecular Medicine (2022)