Abstract
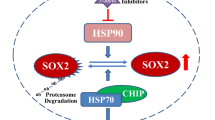
CSN6, a critical subunit of the constitutive photomorphogenesis 9 (COP9) signalosome (CSN), has received attention as a regulator of the degradation of cancer-related proteins such as p53, c-myc and c-Jun, through the ubiquitin-proteasome system, suggesting its importance in cancerogenesis. However, the biological functions and molecular mechanisms of CSN6 in glioblastoma (GBM) remain poorly understood. Here, we report that GBM tumors overexpressed CSN6 compared with normal brain tissues and that CSN6 promoted GBM cell proliferation, migration, invasion and tumorigenesis. Erlotinib, a small-molecule epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, was used to reveal that the proliferative and metastatic effects of CSN6 on GBM cells were EGFR dependent. We also found that CSN6 positively regulated EGFR stability via reduced levels of EGFR ubiquitination, thereby elevating steady expression of EGFR. In addition, this study is the first description of a novel role for the CSN6-interacting E3 ligase, CHIP (carboxyl terminus of heat-shock protein 70-interacting protein), regulating EGFR ubiquitination in cancer cells. We showed that CSN6 associated with CHIP and led to CHIP destabilization by increasing CHIP self-ubiquitination. Moreover, CSN6 decreased CHIP expression and increased EGFR expression in the tumor samples. Deregulation of this axis promoted GBM cell’s proliferation and metastasis. Thus, our study provides insights into the applicability of using the CSN6-CHIP-EGFR axis as a potential therapeutic target in cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol 2002; 61: 215–225; discussion 226–219.
Olar A, Aldape KD . Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol 2014; 232: 165–177.
Riddick G, Fine HA . Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol 2011; 7: 439–450.
Wen PY, Kesari S . Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507.
Wei N, Deng XW . The COP9 signalosome. Annu Rev Cell Dev Biol 2003; 19: 261–286.
Li L, Deng XW . The COP9 signalosome: an alternative lid for the 26S proteasome? Trends Cell Biol 2003; 13: 507–509.
Gusmaroli G, Figueroa P, Serino G, Deng XW . Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis cullin3-based E3 ligases. Plant Cell 2007; 19: 564–581.
Peng Z, Serino G, Deng XW . Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 2001; 13: 2393–2407.
Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001; 292: 1382–1385.
Maytal-Kivity V, Reis N, Hofmann K, Glickman MH . MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem 2002; 3: 28.
Lee MH, Zhao R, Phan L, Yeung SC . Roles of COP9 signalosome in cancer. Cell Cycle 2011; 10: 3057–3066.
Fang L, Lu W, Choi HH, Yeung SC, Tung JY, Hsiao CD et al. ERK2-dependent phosphorylation of CSN6 is critical in colorectal cancer development. Cancer Cell 2015; 28: 183–197.
Gao S, Fang L, Phan LM, Qdaisat A, Yeung SC, Lee MH . COP9 signalosome subunit 6 (CSN6) regulates E6AP/UBE3A in cervical cancer. Oncotarget 2015; 6: 28026–28041.
Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y et al. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res 2000; 60: 4519–4525.
Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature 1985; 313: 144–147.
Yarden Y, Sliwkowski MX . Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2: 127–137.
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21: 2683–2710.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061–1068.
Lipkowitz S . The role of the ubiquitination-proteasome pathway in breast cancer: ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breast Cancer Res 2003; 5: 8–15.
Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 1999; 4: 1029–1040.
Lin DC, Xu L, Chen Y, Yan H, Hazawa M, Doan N et al. Genomic and functional analysis of the E3 ligase PARK2 in glioma. Cancer Res 2015; 75: 1815–1827.
Wang T, Yang J, Xu J, Li J, Cao Z, Zhou L et al. CHIP is a novel tumor suppressor in pancreatic cancer through targeting EGFR. Oncotarget 2014; 5: 1969–1986.
McDonough H, Patterson C . CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 2003; 8: 303–308.
Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J Biol Chem 2012; 287: 15996–16006.
Kajiro M, Hirota R, Nakajima Y, Kawanowa K, So-ma K, Ito I et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol 2009; 11: 312–319.
Cao Z, Xu J, Huang H, Shen P, You L, Zhou L et al. MiR-1178 promotes the proliferation, G1/S transition, migration and invasion of pancreatic cancer cells by targeting CHIP. PLoS One 2015; 10: e0116934.
Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun 2014; 5: 5384.
Ronnebaum SM, Wu Y, McDonough H, Patterson C . The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Mol Cell Biol 2013; 33: 4461–4472.
Downward J, Parker P, Waterfield MD . Autophosphorylation sites on the epidermal growth factor receptor. Nature 1984; 311: 483–485.
Hsuan JJ . Oncogene regulation by growth factors. Anticancer Res 1993; 13: 2521–2532.
Raizer JJ, Abrey LE, Lassman AB, Chang SM, Lamborn KR, Kuhn JG et al. A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro-Oncology 2010; 12: 95–103.
van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol 2009; 27: 1268–1274.
Citri A, Yarden Y . EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7: 505–516.
Xuan F, Huang M, Liu W, Ding H, Yang L, Cui H . Homeobox C9 suppresses Beclin1-mediated autophagy in glioblastoma by directly inhibiting the transcription of death-associated protein kinase 1. Neuro-Oncology 2015; 18: 819–829.
Acknowledgements
This work was supported by the Research Fund for the Doctoral Program of Higher Education of China (20130182110003), the National Basic Research Program of China (No. 2012cb114603) and the National Natural Science Foundation of China (81502574 and 31501100).
Author contributions
JH: Collection and assembly of data, data analysis and interpretation and manuscript writing; QD and JZ: collection and assembly of data and data analysis; JZ, YZ, PT and WZ: collection and assembly of data; HC: concept design, financial support, data analysis and interpretation and manuscript revision.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Hou, J., Deng, Q., Zhou, J. et al. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene 36, 1134–1144 (2017). https://doi.org/10.1038/onc.2016.280
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.280