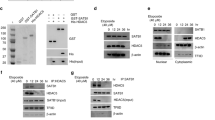
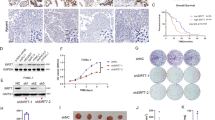
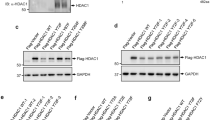
Abstract
Sirt6 is a histone deacetylase with NAD+-dependent activity. Sirt6 has been shown as a tumor suppressor partially via inhibiting the expression of c-Myc target genes and ribosome biogenesis. However, how to regulate Sirt6 activity is largely unknown. In this study, we identify that Sirt6 can be modified by small ubiquitin-like modifier. Sirt6 SUMOylation deficiency specifically decreases its deacetylation of H3K56 but not H3K9 in vivo. Mechanistically, we find that SUMOylation deficiency decreases Sirt6 binding with c-Myc, decreasing Sirt6 occupancy on the locus of c-Myc target genes. Therefore, Sirt6 SUMOylation deficiency reduces its deacetylation of H3k56 and its repression of c-Myc target genes. Moreover, Sirt6 SUMOylation deficiency reduces its suppression of cell proliferation and tumorigenesis. Thus, these results reveal that SUMOylation has an important role in regulation of Sirt6 deacetylation on H3K56, as well as its tumor suppressive activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dutnall RN, Pillus L . Deciphering NAD-dependent deacetylases. Cell 2001; 105: 161–164.
Klar AJ, Strathern JN, Broach JR, Hicks JB . Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature 1981; 289: 239–244.
Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 2011; 477: 482–485.
Chalkiadaki A, Guarente L . Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol 2012; 8: 287–296.
Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R . Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov 2012; 11: 443–461.
Ardestani PM, Liang F . Sub-cellular localization, expression and functions of Sirt6 during the cell cycle in HeLa cells. Nucleus 2012; 3: 442–451.
Bosch-Presegue L, Vaquero A . The dual role of sirtuins in cancer. Genes & cancer 2011; 2: 648–662.
Lombard DB, Schwer B, Alt FW, Mostoslavsky R . SIRT6 in DNA repair, metabolism and ageing. J Intern Med 2008; 263: 128–141.
Tennen RI, Berber E, Chua KF . Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev 2010; 131: 185–192.
Kugel S, Mostoslavsky R . Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci 2014; 39: 72–81.
Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 2009; 136: 62–74.
Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012; 151: 1185–1199.
Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 2013; 51: 454–468.
Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010; 140: 280–293.
Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 2012; 18: 1643–1650.
Kaidi A, Weinert BT, Choudhary C, Jackson SP . Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 2010; 329: 1348–1353.
Dominy Jr JE, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell 2012; 48: 900–913.
Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011; 332: 1443–1446.
Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013; 496: 110–113.
Finkel T, Deng CX, Mostoslavsky R . Recent progress in the biology and physiology of sirtuins. Nature 2009; 460: 587–591.
Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol 2012; 14: 1203–1211.
Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA 2011; 108: 9232–9237.
Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J 2013; 32: 2248–2263.
Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem 2013; 288: 18439–18447.
Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q et al. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep 2013; 5: 1639–1649.
Geiss-Friedlander R, Melchior F . Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8: 947–956.
Gareau JR, Lima CD . The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11: 861–871.
Yeh ET . SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem 2009; 284: 8223–8227.
Cheng J, Kang X, Zhang S, Yeh ET . SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 2007; 131: 584–595.
Takasaka N, Araya J, Hara H, Ito S, Kobayashi K, Kurita Y et al. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Immunol 2014; 192: 958–968.
Tasselli L, Chua KF . Cancer: Metabolism in 'the driver's seat. Nature 2012; 492: 362–363.
Lyssiotis CA, Cantley LC . SIRT6 puts cancer metabolism in the driver's seat. Cell 2012; 151: 1155–1156.
Kawahara TL, Rapicavoli NA, Wu AR, Qu K, Quake SR, Chang HY . Dynamic chromatin localization of Sirt6 shapes stress- and aging-related transcriptional networks. PLoS Genet 2011; 7: e1002153.
Zhang ZG, Qin CY . Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signalregulated kinase signaling pathway. Mol Med Rep 2014; 9: 882–888.
Etchegaray JP, Zhong L, Mostoslavsky R . The histone deacetylase SIRT6: at the crossroads between epigenetics, metabolism and disease. Curr Top Med Chem 2013; 13: 2991–3000.
Gertler AA, Cohen HY . SIRT6, a protein with many faces. Biogerontology 2013; 14: 629–639.
Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y . Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 2004; 101: 14373–14378.
Escobar-Cabrera E, Okon M, Lau DK, Dart CF, Bonvin AM, McIntosh LP . Characterizing the N- and C-terminal Small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J Biol Chem 2011; 286: 19816–19829.
Kolesar P, Sarangi P, Altmannova V, Zhao X, Krejci L . Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res 2012; 40: 7831–7843.
Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell 2010; 38: 191–201.
Acknowledgements
We thank Dr Chuxia Deng (NIH, Bethesda, MD, USA) for kindly providing for Sirt6−/− MEF cells. This work was supported by National Basic Research Program of China (973 Program; no. 2013CB910902 to JC), National Natural Science Foundation of China (91229202 and 81430069 to JC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Cai, J., Zuo, Y., Wang, T. et al. A crucial role of SUMOylation in modulating Sirt6 deacetylation of H3 at lysine 56 and its tumor suppressive activity. Oncogene 35, 4949–4956 (2016). https://doi.org/10.1038/onc.2016.24
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.24
This article is cited by
-
Post-translational modifications of nuclear sirtuins
Genome Instability & Disease (2020)