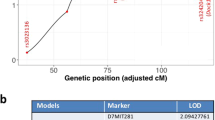
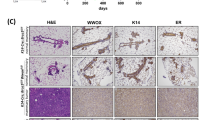
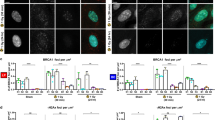
Abstract
Almost half of all hereditary breast cancers (BCs) are associated with germ-line mutations in homologous recombination (HR) genes. However, the tumor phenotypes associated with different HR genes vary, making it difficult to define the role of HR in BC predisposition. To distinguish between HR-dependent and -independent features of BCs, we generated a mouse model in which an essential HR gene, Rad51c, is knocked-out specifically in epidermal tissues. Rad51c is one of the key mediators of HR and a well-known BC predisposition gene. Here, we demonstrate that deletion of Rad51c invariably requires inactivation of the Trp53 tumor suppressor (TP53 in humans) to produce mammary carcinomas in 63% of female mice. Nonetheless, loss of Rad51c shortens the latency of Trp53-deficient mouse tumors from 11 to 6 months. Remarkably, the histopathological features of Rad51c-deficient mammary carcinomas, such as expression of hormone receptors and luminal epithelial markers, faithfully recapitulate the histopathology of human RAD51C-mutated BCs. Similar to other BC models, Rad51c/p53 double-mutant mouse mammary tumors also reveal a propensity for genomic instability, but lack the focal amplification of the Met locus or distinct mutational signatures reported for other HR genes. Using the human mammary epithelial cell line MCF10A, we show that deletion of TP53 can rescue RAD51C-deficient cells from radiation-induced cellular senescence, whereas it exacerbates their centrosome amplification and nuclear abnormalities. Altogether, our data indicate that a trend for genomic instability and inactivation of Trp53 are common features of HR-mediated BCs, whereas histopathology and somatic mutation patterns are specific for different HR genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Olopade OI, Grushko TA, Nanda R, Huo D . Advances in breast cancer: pathways to personalized medicine. Clin Cancer Res 2008; 14: 7988–7999.
Thompson LH, Schild D . Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res - Fundam Mol Mech Mutagen 2001; 477: 131–153.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003; 100: 8418–8423.
Arnes JB, Brunet JS, Stefansson I, Bégin LR, Wong N, Chappuis PO et al. Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 2005; 11: 4003–4011.
Foulkes WD . BRCA1 functions as a breast stem cell regulator. J Med Genet 2004; 41: 1–5.
Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA 2008; 105: 1680–1685.
Munne PM, Gu Y, Tumiati M, Gao P, Koopal S, Uusivirta S et al. TP53 supports basal-like differentiation of mammary epithelial cells by preventing translocation of deltaNp63 into nucleoli. Sci Rep 2014; 4: 4663.
Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A et al. 53BP1 inhibits homologous recombination in brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141: 243–254.
Jaspers JE, Kersbergen A, Boon U, Sol W, Van Deemter L, Zander SA et al. Loss of 53BP1 causes PARP inhibitor resistance in BRCA1-mutated mouse mammary tumors. Cancer Discov 2013; 3: 68–81.
Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle 2012; 11: 3837–3850.
Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 1996; 14: 430–440.
Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T . Transcriptional activation functions in BRCA2. Nature 1997; 386: 772–773.
Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010; 42: 410–414.
Osorio A, Endt D, Fernández F, Eirich K, de la Hoya M, Schmutzler R et al. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet 2012; 21: 2889–2898.
Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 2010; 42: 406–409.
Dosanjh M . Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res 1998; 26: 1179–1184.
Lin Z, Kong H, Nei M, Ma H . Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA 2006; 103: 10328–10333.
Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 2001; 15: 3296–3307.
Chun J, Buechelmaier ES, Powell SN . Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol 2013; 33: 387–395.
Rodrigue A, Coulombe Y, Jacquet K, Gagné J-P, Roques C, Gobeil S et al. The RAD51 paralogs ensure cellular protection against mitotic defects and aneuploidy. J Cell Sci 2013; 126: 348–359.
Somyajit K, Subramanya S, Nagaraju G . Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem 2012; 287: 3366–3380.
Kee Y, D’Andrea AD . Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest 2012; 122: 3799–3806.
Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet 2013; 92: 800–806.
Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 2010; 17: 1461–1469.
Renglin Lindh A, Schultz N, Saleh-Gohari N, Helleday T . RAD51C (RAD51L2) is involved in maintaining centrosome number in mitosis. Cytogenet Genome Res 2007; 116: 38–45.
Katsura M, Tsuruga T, Date O, Yoshihara T, Ishida M, Tomoda Y et al. The ATR-Chk1 pathway plays a role in the generation of centrosome aberrations induced by Rad51C dysfunction. Nucleic Acids Res 2009; 37: 3959–3968.
Badie S, Liao C, Thanasoula M, Barber P, Hill M A, Tarsounas M . RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J Cell Biol 2009; 185: 587–600.
Kee Y, D’Andrea AD . Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 2010; 24: 1680–1694.
Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 2007; 104: 12111–12116.
Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A . Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 2001; 29: 418–425.
Bowman-Colin C, Xia B, Bunting S, Klijn C, Drost R, Bouwman P et al. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc Natl Acad Sci USA 2013; 110: 8632–8637.
Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, Martin BK et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol 2007; 176: 581–592.
Kuznetsov SG, Haines DC, Martin BK, Sharan SK . Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res 2009; 69: 863–872.
Tumiati M, Hemmes A, Uusivirta S, Koopal S, Kankainen M, Lehtonen E et al. Loss of Rad51c accelerates tumourigenesis in sebaceous glands of Trp53-mutant mice. J Pathol 2015; 235: 136–146.
Gevensleben H, Bossung V, Meindl A, Wappenschmidt B, de Gregorio N, Osorio A et al. Pathological features of breast and ovarian cancers in RAD51C germline mutation carriers. Virchows Arch 2014; 465: 365–369.
Knight JF, Lesurf R, Zhao H, Pinnaduwage D, Davis RR, Saleh SMI et al. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc Natl Acad Sci USA 2013; 110: E1301–E1310.
Smolen GA, Muir B, Mohapatra G, Barmettler A, Kim WJ, Rivera MN et al. Frequent met oncogene amplification in a Brca1/Trp53 mouse model of mammary tumorigenesis. Cancer Res 2006; 66: 3452–3455.
Lee JM, Dedhar S, Kalluri R, Thompson EW . The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006; 172: 973–981.
Stratton MR, Rahman N . The emerging landscape of breast cancer susceptibility. Nat Genet 2008; 40: 17–22.
Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 2011; 26: 125–132.
Tacconi EMC, Tarsounas M . How homologous recombination maintains telomere integrity. Chromosoma 2014; 124: 119–130.
Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol 2009; 11: 1135–1142.
Olivares-Illana V, Fåhraeus R . p53 isoforms gain functions. Oncogene 2010; 29: 5113–5119.
Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 1999; 97: 527–538.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 2009; 19: 524–529.
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012; 149: 979–993.
Francis JC, Melchor L, Campbell J, Kendrick H, Wei W, Armisen-Garrido J et al. Whole-exome DNA sequence analysis of Brca2 - and Trp53 -deficient mouse mammary gland tumours. J Pathol 2015; 236: 186–200.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367.
Acknowledgements
We thank Pekka Ellonen, Maija Lepistö, Sonja Lagström, and the rest of the FIMM sequencing unit for advice and sequencing of tumor samples, the pathology unit of the Haartman Institute for histology services, Sami Blom and Dr Johan Lundin for scanning histological slides. We are grateful to Dr Shyam Sharan and Dr Madalena Tarsounas for providing critical comments on the manuscript. Financial support for the project was provided to SGK by the Academy of Finland, Finnish Medical Foundation, Sigrid Juselius Foundation, Cancer Society of Finland, and Biocenter Finland. MT was supported by a fellowship from the Helsinki Doctoral Program in Biomedicine (DPBM), and the Academy of Finland.
Author contributions
MT designed and performed all experiments, analyzed the data, and wrote the manuscript. PMM and AH assisted with animal handling and histology. HE performed CNV analysis. SE analyzed tumor somatic mutations. SGK designed and supervised the project, and wrote the manuscript. All authors reviewed the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Tumiati, M., Munne, P., Edgren, H. et al. Rad51c- and Trp53-double-mutant mouse model reveals common features of homologous recombination-deficient breast cancers. Oncogene 35, 4601–4610 (2016). https://doi.org/10.1038/onc.2015.528
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.528