Abstract
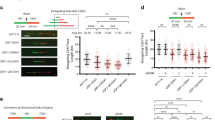
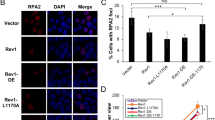
DNA polymerase eta (Polη) has unique and pivotal functions in several DNA damage-tolerance pathways. Steady-state level of this short-lived protein is tightly controlled by multiple mechanisms including proteolysis. Here, we have identified the deubiquitinating enzyme (DUB), ubiquitin-specific protease 7 (USP7), as a novel regulator of Polη stability. USP7 regulates Polη stability through both indirect and direct mechanisms. Knockout of USP7 increased the steady-state level of Polη and slowed down the turnover of both Polη and p53 proteins through destabilizing their E3 ligase murine double minute 2 (Mdm2). Also, USP7 physically binds Polη in vitro and in vivo. Overexpression of wild-type USP7 but not its catalytically-defective mutants deubiquitinates Polη and increases its cellular steady-state level. Thus, USP7 directly serves as a specific DUB for Polη. Furthermore, ectopic expression of USP7 promoted the UV-induced proliferating cell nuclear antigen (PCNA) monoubiquitination in Polη-proficient but not in Polη-deficient XPV (Xeroderma pigmentosum variant) cells, suggesting that USP7 facilitates UV-induced PCNA monoubiquitination by stabilizing Polη. Taken together, our findings reveal a modulatory role of USP7 in PCNA ubiquitination-mediated stress-tolerance pathways by fine-tuning Polη turnover.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Friedberg EC, Lehmann AR, Fuchs RP . Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell 2005; 18: 499–505.
Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007; 6: 891–899.
Durando M, Tateishi S, Vaziri C . A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res 2013; 41: 3079–3093.
Liu G, Chen X . DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol 2006; 26: 1398–1413.
Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 1999; 399: 700–704.
Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M . Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 2004; 23: 3886–3896.
Skoneczna A, McIntyre J, Skoneczny M, Policinska Z, Sledziewska-Gojska E . Polymerase eta is a short-lived, proteasomally degraded protein that is temporarily stabilized following UV irradiation in Saccharomyces cerevisiae. J Mol Biol 2007; 366: 1074–1086.
Kim SH, Michael WM . Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell 2008; 32: 757–766.
Jung YS, Qian Y, Chen X . DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst) 2012; 11: 177–184.
Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N . USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol 2009; 184: 13–19.
He J, Zhu Q, Wani G, Sharma N, Han C, Qian J et al. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem 2014; 289: 27278–27289.
Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8: 339–347.
Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005; 17: 331–339.
Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 2012; 44: 598–602.
Zhang D, Zaugg K, Mak TW, Elledge SJ . A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006; 126: 529–542.
Cummins JM, Vogelstein B . HAUSP is required for p53 destabilization. Cell Cycle 2004; 3: 689–692.
Li M, Brooks CL, Kon N, Gu W . A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004; 13: 879–886.
Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B . Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 2004; 428: 1.
Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci USA 2012; 109: 4828–4833.
Jagannathan M, Nguyen T, Gallo D, Luthra N, Brown GW, Saridakis V et al. A role for USP7 in DNA replication. Mol Cell Biol 2014; 34: 132–145.
Sarasin A . UVSSA and USP7: new players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med 2012; 4: 44.
Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 2012; 44: 593–597.
Acknowledgements
We are thankful to Dr Bert Vogelstein for HCT116, HCT116 p53−/− and HCT116 USP7−/− cells; Dr Alan Lehmann for XP30RO and XP30RO-GFP-Polη cells; Drs Yanhui Xu and Yang Shi for FLAG-tagged WT and CS USP7 constructs; Dr Carol Prives for Myc-Mdm2 construct. This work was supported by grants from the National Institutes of Health (ES012991 and ES023883) to AAW.
Author Contributions
JQ and AAW designed research; JQ, KP and AKS performed research; JQ, KP, QZ, QW, AKS, JH and AAW analyzed data; JQ and AAW wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Qian, J., Pentz, K., Zhu, Q. et al. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 34, 4791–4796 (2015). https://doi.org/10.1038/onc.2014.394
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.394
This article is cited by
-
Ubiquitin specific peptidase 37 and PCNA interaction promotes osteosarcoma pathogenesis by modulating replication fork progression
Journal of Translational Medicine (2023)
-
Ubiquitin-specific peptidase 39 regulates the process of proliferation and migration of human ovarian cancer via p53/p21 pathway and EMT
Medical Oncology (2019)
-
Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases
Signal Transduction and Targeted Therapy (2018)
-
DUB3 and USP7 de-ubiquitinating enzymes control replication inhibitor Geminin: molecular characterization and associations with breast cancer
Oncogene (2017)
-
The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics
Cell & Bioscience (2016)