Abstract
Despite equal snRNP stoichiometry in spliceosomes, U1 snRNP (U1) is typically the most abundant vertebrate snRNP. Mechanisms regulating U1 overabundance and snRNP repertoire are unknown. In Sm-core assembly, a key snRNP-biogenesis step mediated by the SMN complex, the snRNA-specific RNA-binding protein (RBP) Gemin5 delivers pre-snRNAs, which join SMN–Gemin2–recruited Sm proteins. We show that the human U1-specific RBP U1-70K can bridge pre-U1 to SMN–Gemin2–Sm, in a Gemin5-independent manner, thus establishing an additional and U1-exclusive Sm core–assembly pathway. U1-70K hijacks SMN–Gemin2–Sm, enhancing Sm-core assembly on U1s and inhibiting that on other snRNAs, thereby promoting U1 overabundance and regulating snRNP repertoire. SMN–Gemin2's ability to facilitate transactions between different RBPs and RNAs explains its multi-RBP valency and the myriad transcriptome perturbations associated with SMN deficiency in neurodegenerative spinal muscular atrophy. We propose that SMN–Gemin2 is a versatile hub for RNP exchange that functions broadly in RNA metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mount, S.M., Pettersson, I., Hinterberger, M., Karmas, A. & Steitz, J.A. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro . Cell 33, 509–518 (1983).
Will, C.L. & Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011).
Berg, M.G. et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64 (2012).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010).
Vorlová, S. et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell 43, 927–939 (2011).
Almada, A.E., Wu, X., Kriz, A.J., Burge, C.B. & Sharp, P.A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013).
Ntini, E. et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 20, 923–928 (2013).
Baserga, S.J. & Steitz, J.A. The diverse world of small ribonucleoproteins. Cold Spring Harb. Monograph Arch. 24, 359–381 (1993).
Battle, D.J. et al. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 71, 313–320 (2006).
Guthrie, C. & Patterson, B. Spliceosomal snRNAs. Annu. Rev. Genet. 22, 387–419 (1988).
Yong, J., Golembe, T.J., Battle, D.J., Pellizzoni, L. & Dreyfuss, G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 24, 2747–2756 (2004).
Kondo, Y., Oubridge, C., van Roon, A.M. & Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife 4, e04986 (2015).
Leung, A.K., Nagai, K. & Li, J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473, 536–539 (2011).
Pomeranz Krummel, D.A., Oubridge, C., Leung, A.K., Li, J. & Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458, 475–480 (2009).
Raker, V.A., Hartmuth, K., Kastner, B. & Lührmann, R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 19, 6554–6565 (1999).
Urlaub, H., Raker, V.A., Kostka, S. & Lührmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 20, 187–196 (2001).
Weber, G., Trowitzsch, S., Kastner, B., Lührmann, R. & Wahl, M.C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29, 4172–4184 (2010).
Battle, D.J. et al. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 23, 273–279 (2006).
Lau, C.K., Bachorik, J.L. & Dreyfuss, G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 16, 486–491 (2009).
Yong, J., Kasim, M., Bachorik, J.L., Wan, L. & Dreyfuss, G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell 38, 551–562 (2010).
Cauchi, R.J. SMN and Gemins: 'we are family' … or are we?: insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. BioEssays 32, 1077–1089 (2010).
Fischer, U., Englbrecht, C. & Chari, A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2, 718–731 (2011).
Chari, A. et al. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 135, 497–509 (2008).
Grimm, C. et al. Structural basis of assembly chaperone-mediated snRNP formation. Mol. Cell 49, 692–703 (2013).
Zhang, R. et al. Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell 146, 384–395 (2011).
Will, C.L. & Lührmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 (2001).
Matera, A.G. & Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014).
Hamm, J., Dathan, N.A., Scherly, D. & Mattaj, I.W. Multiple domains of U1 snRNA, including U1 specific protein binding sites, are required for splicing. EMBO J. 9, 1237–1244 (1990).
Nelissen, R.L., Will, C.L., van Venrooij, W.J. & Lührmann, R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 13, 4113–4125 (1994).
Yong, J., Pellizzoni, L. & Dreyfuss, G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 21, 1188–1196 (2002).
Wan, L., Ottinger, E., Cho, S. & Dreyfuss, G. Inactivation of the SMN complex by oxidative stress. Mol. Cell 31, 244–254 (2008).
Wan, L. et al. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25, 5543–5551 (2005).
Younis, I. et al. Minor introns are embedded molecular switches regulated by highly unstable U6ata U6atac snRNA. eLife 2, e00780 (2013).
Kohtz, J.D. et al. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124 (1994).
Cléry, A., Blatter, M. & Allain, F.H. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18, 290–298 (2008).
Cho, S. et al. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 108, 8233–8238 (2011).
Burghes, A.H. & Beattie, C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10, 597–609 (2009).
Lorson, C.L. et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 19, 63–66 (1998).
Pellizzoni, L., Yong, J. & Dreyfuss, G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298, 1775–1779 (2002).
Young, P.J. et al. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 9, 2869–2877 (2000).
Bedford, M.T. & Clarke, S.G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 (2009).
Tripsianes, K. et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420 (2011).
Liu, K. et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One 7, e30375 (2012).
Yong, J., Wan, L. & Dreyfuss, G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 14, 226–232 (2004).
Ling, S.C., Polymenidou, M. & Cleveland, D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Piazzon, N. et al. Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle. Nucleic Acids Res. 41, 1255–1272 (2013).
Cheng, D., Côté, J., Shaaban, S. & Bedford, M.T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 (2007).
Lorson, C.L. & Androphy, E.J. The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum. Mol. Genet. 7, 1269–1275 (1998).
O'Reilly, D. et al. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res. 23, 281–291 (2013).
Shukla, S. & Parker, R. Quality control of assembly-defective U1 snRNAs by decapping and 5′-to-3′ exonucleolytic digestion. Proc. Natl. Acad. Sci. USA 111, E3277–E3286 (2014).
Gabanella, F. et al. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2, e921 (2007).
Li, D.K., Tisdale, S., Lotti, F. & Pellizzoni, L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 32, 22–29 (2014).
Tisdale, S. et al. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3′-end formation of histone mRNAs. Cell Reports 5, 1187–1195 (2013).
Zhang, Z. et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 (2008).
Zhang, Z. et al. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 110, 19348–19353 (2013).
Sun, S. et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 6, 6171 (2015).
Tsuiji, H. et al. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol. Med. 5, 221–234 (2013).
Yamazaki, T. et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Reports 2, 799–806 (2012).
Acknowledgements
We thank members of our laboratory for helpful discussions and comments on the manuscript. This work was supported by the Association Française Contre les Myopathies (AFM) and by the US National Institutes of Health (R01 GM112923 to G.D.) G.D. is supported as an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
B.R.S., L.W. and Z.Z. designed and performed experiments. B.R.S., L.W., Z.Z., P.L., E.B., J.D. and I.Y. contributed to data analysis. G.D. is responsible for the project's planning and experimental design. All authors contributed to writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
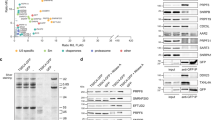
Supplementary Figure 1 Western blot analysis of the Gemin2-knockdown cell extracts used for in vitro Sm-core assembly.
Input cytoplasmic extracts from HeLa cells, transfected with control siRNA and siRNA targeting against Gemin2, are shown. Percent of residual Gemin2 protein after knockdown compared to control (100%) is indicated.
Supplementary Figure 2 Western blot analysis of the U1-70K-, SMN- or Gemin5-knockdown cell extracts used for snRNP-level measurements.
The SMN complex, U1-70K and Sm proteins in control, U1-70K, SMN or Gemin5 knockdown HeLa cells are shown. FXR1 was used as a loading control. Residual knockdown proteins for each knockdown compared to control (100%) are indicated.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 8152 kb)
Supplementary Data Set 1
Uncropped blots and gels with size marker indications (PDF 8111 kb)
Rights and permissions
About this article
Cite this article
So, B., Wan, L., Zhang, Z. et al. A U1 snRNP–specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23, 225–230 (2016). https://doi.org/10.1038/nsmb.3167
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3167
This article is cited by
-
The SMN complex drives structural changes in human snRNAs to enable snRNP assembly
Nature Communications (2023)
-
Phylogenetic comparison and splice site conservation of eukaryotic U1 snRNP-specific U1-70K gene family
Scientific Reports (2021)
-
Hyper-SUMOylation of SMN induced by SENP2 deficiency decreases its stability and leads to spinal muscular atrophy-like pathology
Journal of Molecular Medicine (2021)
-
Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder
Nature Communications (2021)
-
Chromatin-associated RNAs as facilitators of functional genomic interactions
Nature Reviews Genetics (2019)