Key Points
-
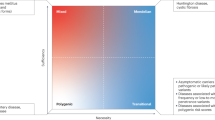
Genome-wide association studies of rheumatic diseases have revealed different shared 'genetic architectures' between seropositive and the seronegative rheumatic diseases
-
SNPs in or near BLK, IRF5, PTPN22, STAT4 and HLA class II genes are associated with many seropositive rheumatic diseases
-
Variants in or near IL23R, IL12B, ERAP1, and HLA class I genes are associated with many seronegative diseases; the ERAP1 association is limited to individuals with the disease-associated HLA class I type
-
Technological advancements in collection and analysis of high-density genetic data could lead to personalized treatment of rheumatic diseases
Abstract
Rheumatoid arthritis, systemic lupus erythematosus, ankylosing spondylitis and some other rheumatic diseases are genetically complex, with evidence of familial clustering, but not of Mendelian inheritance. These diseases are thought to result from contributions and interactions of multiple genetic and nongenetic risk factors, which have small effects individually. Genome-wide association studies (GWAS) of large collections of data from cases and controls have revealed many genetic factors that contribute to non-Mendelian rheumatic diseases, thus providing insights into associated molecular mechanisms. This Review summarizes methods for the identification of gene variants that influence genetically complex diseases and focuses on what we have learned about the rheumatic diseases for which GWAS have been reported. Our review of the disease-associated loci identified to date reveals greater sharing of risk loci among the groups of seropositive (diseases in which specific autoantibodies are often present) or seronegative diseases than between these two groups. The nature of the shared and discordant loci suggests important similarities and differences among these diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mahdi, H. et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated α-enolase in the etiology of rheumatoid arthritis. Nat. Genet. 41, 1319–1324 (2009).
Costenbader, K. H., Gay, S., Alarcon-Riquelme, M. E., Iaccarino, L. & Doria, A. Genes, epigenetic regulation and environmental factors: which is the most relevant in developing autoimmune diseases? Autoimmun. Rev. 11, 604–609 (2012).
Honda, K. & Littman, D. R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30, 759–795 (2012).
Manolio, T. A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363, 166–176 (2010).
Heliovaara, M., Aho, K., Aromaa, A., Knekt, P. & Reunanen, A. Smoking and risk of rheumatoid arthritis. J. Rheumatol. 20, 1830–1835 (1993).
Katz, U. & Zandman-Goddard, G. Drug-induced lupus: an update. Autoimmun. Rev. 10, 46–50 (2010).
Chen, M., Gao, Y., Guo, X. H. & Zhao, M. H. Propylthiouracil-induced antineutrophil cytoplasmic antibody-associated vasculitis. Nat. Rev. Nephrol. 8, 476–483 (2012).
Fumagalli, M. et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 7, e1002355 (2011).
Raj, T. et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am. J. Hum. Genet. 92, 517–529 (2013).
Lander, E. S. The new genomics: global views of biology. Science 274, 536–539 (1996).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22, 231–238 (1999).
International HapMap Consortium. International HapMap project [online], (2015).
1000 Genomes. A deep catalog of human genetic variation [online], (2015).
International HapMap Consortium. The international HapMap project. Nature 426, 789–796 (2003).
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Kathiresan, S. et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197 (2008).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Stahl, E. A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Okada, Y. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 44, 511–516 (2012).
Kim, K. et al. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann. Rheum. Dis. 74, e13 (2015).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013).
Dilthey, A. T., Moutsianas, L., Leslie, S. & McVean, G. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27, 968–972 (2011).
Pyo, C. W. et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics 14, 89 (2013).
National Human Genome Research Institute. A catalog of published genome-wide association studies. National Institutes of Health [online], (2015).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Onouchi, Y. et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat. Genet. 44, 517–521 (2012).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987).
Reveille, J. D. et al. Systemic lupus erythematosus in three ethnic groups: I. The effects of HLA class II, C4, and CR1 alleles, socioeconomic factors, and ethnicity at disease onset. LUMINA study group. Lupus in minority populations, nature versus nurture. Arthritis Rheum. 41, 1161–1172 (1998).
Gorlova, O. et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS Genet. 7, e1002178 (2011).
Lyons, P. A. et al. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 367, 214–223 (2012).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Mayes, M. D. et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am. J. Hum. Genet. 94, 47–61 (2014).
Morris, D. L. et al. Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am. J. Hum. Genet. 91, 778–793 (2012).
Saruhan-Direskeneli, G. et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am. J. Hum. Genet. 93, 298–305 (2013).
Terao, C. et al. Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the IL12B and HLA-B regions in a Japanese population. Am. J. Hum. Genet. 93, 289–297 (2013).
Remmers, E. F. et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet's disease. Nat. Genet. 42, 698–702 (2010).
Evans, D. M. et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 (2011).
Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Palm, O., Moum, B., Ongre, A. & Gran, J. T. Prevalence of ankylosing spondylitis and other spondyloarthropathies among patients with inflammatory bowel disease: a population study (the IBSEN study). J. Rheumatol. 29, 511–515 (2002).
Bonfiglioli, R. et al. Frequency of HLA-B27 alleles in Brazilian patients with psoriatic arthritis. Clin. Rheumatol. 27, 709–712 (2008).
Bowness, P. et al. TH17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J. Immunol. 186, 2672–2680 (2011).
Ombrello, M. J. et al. Behcet disease-associated MHC class I residues implicate antigen binding and regulation of cell-mediated cytotoxicity. Proc. Natl Acad. Sci. USA 111, 8867–8872 (2014).
Okada, Y. et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 95, 162–172 (2014).
Achkar, J. P. et al. Amino acid position 11 of HLA-DRβ1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 13, 245–252 (2012).
Quinton, J. F. et al. Anti-saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut 42, 788–791 (1998).
Padyukov, L. et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis. 70, 259–265 (2011).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Han, B. et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am. J. Hum. Genet. 94, 522–532 (2014).
Bossini-Castillo, L. et al. A genome-wide association study of rheumatoid arthritis without antibodies against citrullinated peptides. Ann. Rheum. Dis. 74, e15 (2014).
Irigoyen, P. et al. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum. 52, 3813–3818 (2005).
Verpoort, K. N. et al. Association of HLA-DR3 with anti-cyclic citrullinated peptide antibody-negative rheumatoid arthritis. Arthritis Rheum. 52, 3058–3062 (2005).
Juji, T., Satake, M., Honda, Y. & Doi, Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens 24, 316–319 (1984).
Haines, J. L. et al. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. The Multiple Sclerosis Genetics Group. Nat. Genet. 13, 469–471 (1996).
Fraussen, J., Claes, N., de Bock, L. & Somers, V. Targets of the humoral autoimmune response in multiple sclerosis. Autoimmun. Rev. 13, 1126–1137 (2014).
Baccala, R., Hoebe, K., Kono, D. H., Beutler, B. & Theofilopoulos, A. N. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat. Med. 13, 543–551 (2007).
Hall, J. C. & Rosen, A. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat. Rev. Rheumatol. 6, 40–49 (2010).
Kirino, Y. et al. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 (2013).
Haroon, N. & Inman, R. D. Endoplasmic reticulum aminopeptidases: biology and pathogenic potential. Nat. Rev. Rheumatol. 6, 461–467 (2010).
Reeves, E., Edwards, C. J., Elliott, T. & James, E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J. Immunol. 191, 35–43 (2013).
York, I. A., Brehm, M. A., Zendzian, S., Towne, C. F. & Rock, K. L. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc. Natl Acad. Sci. USA 103, 9202–9207 (2006).
Di Cesare, A., Di Meglio, P. & Nestle, F. O. The IL-23/TH17 axis in the immunopathogenesis of psoriasis. J. Invest. Dermatol. 129, 1339–1350 (2009).
Sherlock, J. P. et al. IL-23 induces spondyloarthropathy by acting on ROR-γt+ CD3+CD4−CD8− entheseal resident T cells. Nat. Med. 18, 1069–1076 (2012).
Suzuki, A. et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34, 395–402 (2003).
Zaitlen, N. et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 9, e1003520 (2013).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Stahl, E. A. et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44, 483–489 (2012).
Casanova, J. L. & Abel, L. Revisiting Crohn's disease as a primary immunodeficiency of macrophages. J. Exp. Med. 206, 1839–1843 (2009).
Panoutsopoulou, K., Tachmazidou, I. & Zeggini, E. In search of low-frequency and rare variants affecting complex traits. Hum. Mol. Genet. 22, R16–21 (2013).
Zuk, O. et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl Acad. Sci. USA 111, E455–E464 (2014).
Diogo, D. et al. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am. J. Hum. Genet. 92, 15–27 (2013).
Kirino, Y. et al. Targeted resequencing implicates the familial Mediterranean fever gene MEFV and the Toll-like receptor 4 gene TLR4 in Behcet disease. Proc. Natl Acad. Sci. USA 110, 8134–8139 (2013).
Hunt, K. A. et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 498, 232–235 (2013).
Tang, H. et al. A large-scale screen for coding variants predisposing to psoriasis. Nat. Genet. 46, 45–50 (2014).
Henrichsen, C. N., Chaignat, E. & Reymond, A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 18, R1–R8 (2009).
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Belot, A. et al. Protein kinase cδ deficiency causes mendelian systemic lupus erythematosus with B cell-defective apoptosis and hyperproliferation. Arthritis Rheum. 65, 2161–2171 (2013).
Okada, Y. et al. Integration of sequence data from a Consanguineous family with genetic data from an outbred population identifies PLB1 as a candidate rheumatoid arthritis risk gene. PLoS ONE 9, e87645 (2014).
Zhou, Q. et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. N. Engl. J. Med. 370, 911–920 (2014).
Navon Elkan, P. et al. Mutant adenosine deaminase 2 in a polyarteritis nodosa vasculopathy. N. Engl. J. Med. 370, 921–931 (2014).
Tanaka, N. et al. High incidence of NLRP3 somatic mosaicism in patients with chronic infantile neurologic, cutaneous, articular syndrome: results of an international multicenter collaborative Study. Arthritis Rheum. 63, 3625–3632 (2011).
Izawa, K. et al. Detection of base substitution-type somatic mosaicism of the NLRP3 gene with >99.9% statistical confidence by massively parallel sequencing. DNA Res. 19, 143–152 (2012).
Omoyinmi, E. et al. Brief report: whole-exome sequencing revealing somatic NLRP3 mosaicism in a patient with chronic infantile neurologic, cutaneous, articular syndrome. Arthritis Rheumatol. 66, 197–202 (2014).
Holzelova, E. et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 351, 1409–1418 (2004).
Lou, D. I. et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl Acad. Sci. USA 110, 19872–19877 (2013).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Ombrello, M. J., Sikora, K. A. & Kastner, D. L. Genetics, genomics, and their relevance to pathology and therapy. Best Pract. Res. Clin. Rheumatol. 28, 175–189 (2014).
Mease, P. et al. Abatacept in the treatment of patients with psoriatic arthritis: results of a six-month, multicenter, randomized, double-blind, placebo-controlled, phase II trial. Arthritis Rheum. 63, 939–948 (2011).
de Paoli, F., Nielsen, B., Rasmussen, F., Deleuran, B. & Sondergaard, K. Abatacept induces clinical improvement in patients with severe systemic sclerosis. Scand. J. Rheumatol 43, 342–345 (2014).
Jostins, L. & Barrett, J. C. Genetic risk prediction in complex disease. Hum. Mol. Genet. 20, R182–R188 (2011).
Xie, G. et al. Association of granulomatosis with polyangiitis (Wegener's) with HLA-DPB1*04 and SEMA6A gene variants: evidence from genome-wide analysis. Arthritis Rheum. 65, 2457–2468 (2013).
Khor, C. C. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 43, 1241–1246 (2011).
Lee, Y. C. et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 44, 522–525 (2012).
Lessard, C. J. et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren's syndrome. Nat. Genet. 45, 1284–1292 (2013).
Li, Y. et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjögren's syndrome at 7q11.23. Nat. Genet. 45, 1361–1365 (2013).
Radstake, T. R. et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 42, 426–429 (2010).
Allanore, Y. et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 7, e1002091 (2011).
Coustet, B. et al. C8orf13-BLK is a genetic risk locus for systemic sclerosis and has additive effects with BANK1: results from a large french cohort and meta-analysis. Arthritis Rheum. 63, 2091–2096 (2011).
Martin, J. E. et al. Identification of CSK as a systemic sclerosis genetic risk factor through genome wide association study follow-up. Hum. Mol. Genet. 21, 2825–2835 (2012).
Zochling, J. et al. An Immunochip based interrogation of scleroderma susceptibility variants identifies a novel association at DNASE1L3. Arthritis Res. Ther. 16, 438 (2014).
Remmers, E. F. et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med. 357, 977–986 (2007).
Yang, W. et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 92, 41–51 (2013).
Okada, Y. et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 8, e1002455 (2012).
Yang, J. et al. ELF1 is associated with systemic lupus erythematosus in Asian populations. Hum. Mol. Genet. 20, 601–607 (2011).
Han, J. W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Kozyrev, S. V. et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 40, 211–216 (2008).
Hom, G. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 358, 900–909 (2008).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233 (2009).
Harley, J. B. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 40, 204–210 (2008).
Manjarrez-Orduno, N. et al. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat. Genet. 44, 1227–1230 (2012).
Jacob, C. O. et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc. Natl Acad. Sci. USA 106, 6256–6261 (2009).
Lessard, C. J. et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am. J. Hum. Genet. 90, 648–660 (2012).
Armstrong, D. L. et al. GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun. 15, 347–354 (2014).
Ramos, P. S. et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 7, e1002406 (2011).
Cunninghame Graham, D. S. et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 7, e1002341 (2011).
Ramos, P. S. et al. Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 63, 2049–2057 (2011).
Kim-Howard, X. et al. Allelic heterogeneity in NCF2 associated with systemic lupus erythematosus (SLE) susceptibility across four ethnic populations. Hum. Mol. Genet. 23, 1656–1668 (2014).
Molineros, J. E. et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 9, e1003222 (2013).
Kim, K. et al. Variation in the ICAM1-ICAM4-ICAM5 locus is associated with systemic lupus erythematosus susceptibility in multiple ancestries. Ann. Rheum. Dis. 71, 1809–1814 (2012).
Yang, W. et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 6, e1000841 (2010).
Sheng, Y. J. et al. Follow-up study identifies two novel susceptibility loci PRKCB and 8p11.21 for systemic lupus erythematosus. Rheumatology (Oxford) 50, 682–688 (2011).
Zhang, J. et al. Three SNPs in chromosome 11q23.3 are independently associated with systemic lupus erythematosus in Asians. Hum. Mol. Genet. 23, 524–533 (2014).
Zhang, Y. et al. Meta-analysis of GWAS on two Chinese populations followed by replication identifies novel genetic variants on the X chromosome associated with systemic lupus erythematosus. Hum. Mol. Genet. 24, 274–285 (2015).
Yu, Z. Y. et al. One novel susceptibility locus associate with systemic lupus erythematosus in Chinese Han population. Rheumatol. Int. 33, 2079–2083 (2013).
Deng, Y. et al. MicroRNA-3148 modulates allelic expression of Toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 9, e1003336 (2013).
Hughes, T. et al. Fine-mapping and transethnic genotyping establish IL2/IL21 genetic association with lupus and localize this genetic effect to IL21. Arthritis Rheum. 63, 1689–1697 (2011).
Hinks, A. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013).
Raychaudhuri, S. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 41, 1313–1318 (2009).
Raychaudhuri, S. et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 40, 1216–1223 (2008).
Suzuki, A. et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat. Genet. 40, 1224–1229 (2008).
Jiang, L. et al. Novel risk Loci for rheumatoid arthritis in Han Chinese and congruence with risk variants in Europeans. Arthritis Rheumatol. 66, 1121–1132 (2014).
McAllister, K. et al. Identification of BACH2 and RAD51B as rheumatoid arthritis susceptibility loci in a meta-analysis of genome-wide data. Arthritis Rheum. 65, 3058–3062 (2013).
Teruel, M. et al. Association of CD247 polymorphisms with rheumatoid arthritis: a replication study and a meta-analysis. PLoS ONE 8, e68295 (2013).
Myouzen, K. et al. Functional variants in NFKBIE and RTKN2 involved in activation of the NF-kB pathway are associated with rheumatoid arthritis in Japanese. PLoS Genet. 8, e1002949 (2012).
Terao, C. et al. Myelin basic protein as a novel genetic risk factor in rheumatoid arthritis—a genome-wide study combined with immunological analyses. PLoS ONE 6, e20457 (2011).
Terao, C. et al. The human AIRE gene at chromosome 21q22 is a genetic determinant for the predisposition to rheumatoid arthritis in Japanese population. Hum. Mol. Genet. 20, 2680–2685 (2011).
Kochi, Y. et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet. 42, 515–519 (2010).
Anderson, C. A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252 (2011).
Yang, S. K. et al. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63, 80–87 (2014).
Yamazaki, K. et al. A genome-wide association study identifies 2 susceptibility Loci for Crohn's disease in a Japanese population. Gastroenterology 144, 781–788 (2013).
Julia, A. et al. A genome-wide association study on a southern European population identifies a new Crohn's disease susceptibility locus at RBX1-EP300. Gut 62, 1440–1445 (2013).
Kenny, E. E. et al. A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci. PLoS Genet. 8, e1002559 (2012).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Cortes, A. et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
Lin, Z. et al. A genome-wide association study in Han Chinese identifies new susceptibility loci for ankylosing spondylitis. Nat. Genet. 44, 73–77 (2012).
Reveille, J. D. et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 42, 123–127 (2010).
Capon, F. et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum. Mol. Genet. 17, 1938–1945 (2008).
Sun, L. D. et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat. Genet. 42, 1005–1009 (2010).
Nair, R. P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Ellinghaus, E. et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat. Genet. 42, 991–995 (2010).
Stuart, P. E. et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat. Genet. 42, 1000–1004 (2010).
Cheng, H. et al. Identification of a missense variant in LNPEP that confers psoriasis risk. J. Invest. Dermatol. 134, 359–365 (2014).
Tsoi, L. C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
Ellinghaus, E. et al. Genome-wide meta-analysis of psoriatic arthritis identifies susceptibility locus at REL. J. Invest. Dermatol. 132, 1133–1140 (2012).
Apel, M. et al. Variants in RUNX3 contribute to susceptibility to psoriatic arthritis, exhibiting further common ground with ankylosing spondylitis. Arthritis Rheum. 65, 1224–1231 (2013).
Huffmeier, U. et al. Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat. Genet. 42, 996–999 (2010).
Mizuki, N. et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet's disease susceptibility loci. Nat. Genet. 42, 703–706 (2010).
Li, H. et al. TNFAIP3 gene polymorphisms confer risk for Behcet's disease in a Chinese Han population. Hum. Genet. 132, 293–300 (2013).
Xavier, J. M. et al. FUT2: filling the gap between genes and environment in Behcet's disease? Ann. Rheum. Dis. 74, 618–624 (2013).
Saruhan-Direskeneli, G. et al. Identification of multiple genetic susceptibility loci in Takayasu arteritis. Am. J. Hum. Genet. 93, 298–305 (2013).
Edwards, S. L., Beesley, J., French, J. D. & Dunning, A. M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 (2013).
Musunuru, K. et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–9 (2010).
The Broad Institute [online], (2015).
ENCODE. Encyclopedia of DNA elements [online], (2015).
National Institutes of Health. Roadmap epigenomics project [online], (2015).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Hu, X. et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet. 89, 496–506 (2011).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Acknowledgements
The authors thank M. J. Ombrello and D. L. Kastner for their comments on this manuscript. Y.K. is supported by grants from Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Grant No. 26713036), The Naito Memorial Foundation, The Uehara Memorial Foundation, Japan Intractable Diseases Research Foundation, Japan Rheumatic Disease Foundation, and Yokohama Foundation for Advancement of Medical Science.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to researching data for the article, providing substantial contribution to discussions of the content, to writing the article and to review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kirino, Y., Remmers, E. Genetic architectures of seropositive and seronegative rheumatic diseases. Nat Rev Rheumatol 11, 401–414 (2015). https://doi.org/10.1038/nrrheum.2015.41
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2015.41
This article is cited by
-
Biological classification of childhood arthritis: roadmap to a molecular nomenclature
Nature Reviews Rheumatology (2021)
-
A high-resolution HLA imputation system for the Taiwanese population: a study of the Taiwan Biobank
The Pharmacogenomics Journal (2020)
-
Nonendocrine mechanisms of sex bias in rheumatic diseases
Nature Reviews Rheumatology (2019)
-
GWAS-identified CCR1 and IL10 loci contribute to M1 macrophage-predominant inflammation in Behçet’s disease
Arthritis Research & Therapy (2018)
-
What rheumatologists need to know about CRISPR/Cas9
Nature Reviews Rheumatology (2017)