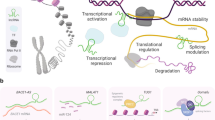
Key Points
-
Although most of the non-coding RNA (ncRNA) species were initially dismissed as products of spurious transcription, a wide spectrum of ncRNA regulatory mechanisms is now emerging.
-
ncRNA expression in the brain is dynamically regulated in an activity-dependent and spatiotemporally controlled manner, suggesting that ncRNAs have precise regulatory roles in brain development and function.
-
The intricate transcriptional output of genomic loci may have affected human brain evolution and could possibly explain the specific vulnerability of the human brain to neurodegeneration.
-
ncRNA expression and function is perturbed in neurodegenerative disorders, and genetic variations in ncRNA networks can be associated with disease risk.
-
Understanding the mechanistic aspects of ncRNA function in the CNS and how ncRNA dysfunction may lead to neurodegenerative disorders will probably lead to the development of new diagnostic and therapeutic approaches for these diseases.
Abstract
The emerging complexity of the transcriptional landscape poses great challenges to our conventional preconceptions of how the genome regulates brain function and dysfunction. Non-protein-coding RNAs (ncRNAs) confer a high level of intricate and dynamic regulation of various molecular processes in the CNS and they have been implicated in neurodevelopment and brain ageing, as well as in synapse function and cognitive performance, in both health and disease. ncRNA-mediated processes may be involved in various aspects of the pathogenesis of neurodegenerative disorders. Understanding these events may help to develop novel diagnostic and therapeutic tools. Here, we provide an overview of the complex mechanisms that are affected by the diverse ncRNA classes that have been implicated in neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Morris, K. V. & Mattick, J. S. The rise of regulatory RNA. Nat. Rev. Genet. 15, 423–437 (2014).
Kadakkuzha, B. M. et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. https://doi.org/10.3389/fncel.2015.00063 (2015).
Webb, A. et al. RNA sequencing of transcriptomes in human brain regions: protein-coding and non-coding RNAs, isoforms and alleles. BMC Genomics 16, 990 (2015).
Salta, E. & De Strooper, B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 11, 189–200 (2012).
Smalheiser, N. R. et al. Natural antisense transcripts are co-expressed with sense mRNAs in synaptoneurosomes of adult mouse forebrain. Neurosci. Res. 62, 236–239 (2008).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015). One of the first reports on circRNA functionality in the mammalian brain.
You, X. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610 (2015). One of the first reports on the role of circRNA in synaptic plasticity.
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Harrow, J. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Mehler, M. F. & Mattick, J. S. Non-coding RNAs in the nervous system. J. Physiol. 5752, 333–341 (2006).
Qureshi, I. A. & Mehler, M. F. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 10, 632–646 (2013).
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L. & Barry, G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877 (2015).
Mercer, T. R. et al. Noncoding RNAs in long-term memory formation. Neuroscientist 14, 434–445 (2008).
Bernard, D. et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093 (2010).
Rajasethupathy, P. et al. A role for neuronal piRNAs in the epigenetic control of memory- related synaptic plasticity. Cell 149, 693–707 (2012).
Lin, N. et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019 (2014).
Bond, A. M. et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027 (2009).
Onoguchi, M. M., Hirabayashi, Y. Y., Koseki, H. H. & Gotoh, Y. Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc. Natl Acad. Sci. USA 109, 16939–16944 (2012).
Ng, S.-Y., Johnson, R. & Stanton, L. W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533 (2012).
Mercer, T. R. et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11, 14 (2010).
Tochitani, S. & Hayashizaki, Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 372, 691–696 (2008).
Rani, N. et al. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188 (2016).
Sauvageau, M. et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 1–24 (2013). This paper provides a solid in vivo proof of concept for the functionality of lincRNAs in the CNS.
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2014).
Hébert, S. S. et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19, 3959–3969 (2010). This is the first paper to demonstrate a causal link between miRNAs and hippocampal neurodegeneration.
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Goff, L. A. et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 112, 6855–6862 (2015).
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F. & Mattick, J. S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–721 (2008). This is the first report on the specificity of the expression of lncRNAs in the mammalian brain.
Belgard, T. G. et al. A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616 (2011).
Lipovich, L. et al. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 192, 1133–1148 (2012).
Kim, T. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Smalheiser, N. R., Lugli, G., Thimmapuram, J., Cook, E. H. & Larson, J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA 17, 166–181 (2011).
Mukilan, M., Ragu Varman, D., Sudhakar, S. & Rajan, K. E. Activity-dependent expression of miR-132 regulates immediate-early gene induction during olfactory learning in the greater short-nosed fruit bat, Cynopterus sphinx. Neurobiol. Learn. Mem. 120, 41–51 (2015).
Barry, G. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494 (2013).
Kumar, L., Shamsuzzama Haque, R., Baghel, T. & Nazir, A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol. Neurobiol. http://dx.doi.org/10.1007/s12035-016-0213-8 (2016).
Liu, S. J. et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17, 67 (2016). First single-cell analysis of ncRNAs in the human brain.
Ayana, R., Singh, S. & Pati, S. Decoding crucial LncRNAs implicated in neurogenesis and neurological disorders. Stem Cells Dev. 26, 541–553 (2017).
Ramos, A. D., Attenello, F. J. & Lim, D. A. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 625, 70–79 (2016).
Ng, S.-Y., Lin, L., Soh, B. S. & Stanton, L. W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 29, 461–468 (2013).
Roberts, T. C., Morris, K. V. & Wood, M. J. A. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Phil. Trans. R. Soc. B Biol. Sci. 369, 20130507 (2014).
Davis, G. M., Haas, M. A. & Pocock, R. MicroRNAs: not 'fine-tuners' but key regulators of neuronal development and function. Front. Neurol. https://doi.org/10.3389/fneur.2015.00245 (2015).
Salta, E. et al. A self-organizing miR-132/Ctbp2 circuit regulates bimodal notch signals and glial progenitor fate choice during spinal cord maturation. Dev. Cell 30, 423–436 (2014).
Sun, A. X., Crabtree, G. R. & Yoo, A. S. MicroRNAs: regulators of neuronal fate. Curr. Opin. Cell Biol. 25, 215–221 (2013).
Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K. & Gage, F. H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793 (2004).
Li, J.-S. & Yao, Z.-X. MicroRNAs: novel regulators of oligodendrocyte differentiation and potential therapeutic targets in demyelination-related diseases. Mol. Neurobiol. 45, 200–212 (2012).
Coolen, M. & Bally-Cuif, L. MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 19, 461–470 (2009).
Weiß, K., Antoniou, A. & Schratt, G. Non-coding mechanisms of local mRNA translation in neuronal dendrites. Eur. J. Cell Biol. http://dx.doi.org/10.1016/j.ejcb.2015.05.011 (2015).
Zhao, X. et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16, 1024–1031 (2013).
Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459 (2012). This paper is one of the first reports on endogenous regulation by NATs in the brain and is the first to coin the term 'antagoNATs' for antisense therapeutic targeting of endogenous NATs.
Maag, J. L. V. et al. Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. http://dx.doi.org/10.3389/fnins.2015.00351 (2015).
Bicker, S., Lackinger, M., Weiß, K. & Schratt, G. MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell. Mol. Life Sci. 71, 3987–4005 (2014).
Schratt, G. microRNAs at the synapse. Nat. Rev. Neurosci. 10, 842–849 (2009).
Smalheiser, N. R. The RNA-centred view of the synapse: non-coding RNAs and synaptic plasticity. Phil. Trans. R. Soc. B Biol. Sci. 369, 20130504 (2014).
Alberini, C. M. & Kandel, E. R. The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 7, a021741 (2015).
Qureshi, I. A. & Mehler, M. F. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol. Med. 17, 337–346 (2011).
Meng, L. et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412 (2014). This is the first paper to demonstrate the potential of therapeutic targeting of lncRNAs in neurological disorders.
St. Laurent, G. & Wahlestedt, C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 30, 612–621 (2007).
Cooper-Knock, J., Kirby, J., Highley, R. & Shaw, P. J. The spectrum of C9orf72-mediated neurodegeneration and amyotrophic lateral sclerosis. Neurotherapeutics 12, 326–339 (2015).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). This is one of the first reports on the causal link between ncRNA expansion repeats and FTD and ALS.
Lagier-Tourenne, C. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl Acad. Sci. USA 110, E4530–E4539 (2013). This paper demonstrates the therapeutic potential of using ASOs against coding and noncoding RNA in ALS and FTD.
Zu, T. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013). This is one of the first papers on the complexity of the C9ORF72 locus.
Taylor, J. P., Brown, R. H. & Cleveland, D. W. Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016).
Moseley, M. L. et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 38, 758–769 (2006). This is the first paper to systematically describe the transcriptional complexity of SCA8.
Nemes, J. P., Benzow, K. A., Moseley, M. L., Ranum, L. P. & Koob, M. D. The SCA8 transcript is an antisense RNA to a brain-specific transcript encoding a novel actin-binding protein (KLHL1). Hum. Mol. Genet. 9, 1543–1551 (2000).
Pastori, C. et al. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 133, 59–67 (2014)
Ladd, P. D. et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187 (2007). This is the first report of ncRNAs in FXS.
Bassell, G. J. & Warren, S. T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214 (2008).
Khalil, A. M., Faghihi, M. A., Modarresi, F., Brothers, S. P. & Wahlestedt, C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE 3, e1486 (2008).
Peschansky, V. J. et al. The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol. Cell. Neurosci. 74, 49–57 (2016).
Bañez-Coronel, M. et al. A pathogenic mechanism in huntington's disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 8, e1002481 (2012).
Chung, D. W., Rudnicki, D. D., Yu, L. & Margolis, R. L. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477 (2011). This is the first study to identify an HD-related ncRNA.
Kordasiewicz, H. B. et al. Sustained therapeutic reversal of huntington's disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044 (2012).
Burd, C. E. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, 1–15 (2010).
Popov, N. & Gil, J. Epigenetic regulation of the INK4B-ARF-INK4a locus: in sickness and in health. Epigenetics 5, 685–690 (2010).
Züchner, S. et al. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann. Hum. Genet. 72, 725–731 (2008).
Emanuele, E. et al. Chromosome 9p21.3 genotype is associated with vascular dementia and Alzheimer's disease. Neurobiol. Aging 32, 1231–1235 (2011).
Arendt, T., Holzer, M. & Gärtner, U. Neuronal expression of cycline dependent kinase inhibitors of the INK4 family in Alzheimer's disease. J. Neural Transm. (Vienna) 105, 949–960 (1998).
Airavaara, M. et al. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 286, 45093–45102 (2011).
Sopher, B. L. et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron 70, 1071–1084 (2011).
Johnson, R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 46, 245–254 (2012).
Xie, Y., Hayden, M. R. & Xu, B. BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J. Neurosci. 30, 14708–14718 (2010).
Chakravarty, D. et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 (2014).
Ferrer, I., Goutan, E., Marín, C., Rey, M. J. & Ribalta, T. Brain-derived neurotrophic factor in Huntington disease. Brain Res. 866, 257–261 (2000).
Xu, L., Zhang, Z., Xie, T., Zhang, X. & Dai, T. Inhibition of BDNF-AS provides neuroprotection for retinal ganglion cells against ischemic injury. PLoS ONE 11, e0164941 (2016).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Santa-Maria, I. et al. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Invest. 125, 681–686 (2015).
Lehmann, S. M. et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835 (2012).
Roshan, R. et al. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA 20, 1287–1297 (2014).
Salta, E. & De Strooper, B. microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer's disease. FASEB J. 31, 424–433 (2017).
Lau, P. et al. Alteration of the microRNA network during the progression of Alzheimer's disease. EMBO Mol. Med 5, 1613–1634 (2013). This study provides a systematic and comprehensive analysis of microRNA expression profiles in the human AD brain.
Salta, E., Sierksma, A., Eynden, Vanden, E. & De Strooper, B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer' s brain. EMBO Mol. Med. 8, 1005–1018 (2016).
Wong, H. K. A. et al. De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer's disease. Hum. Mol. Genet. 22, 3077–3092 (2013).
Ciarlo, E. et al. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis. Model. Mech. 6, 424–433 (2013).
Andersen, O. M. et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry 45, 2618–2628 (2006).
Massone, S. et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317 (2011).
Daughters, R. S. et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 5, e1000600 (2009).
Faghihi, M. A. & Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10, 637–643 (2009).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of ß-secretase. Nat. Med. 14, 723–730 (2008).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense. RNA. EMBO J. 30, 4414–4422 (2011).
Zhao, Y., Alexandrov, P., Jaber, V. & Lukiw, W. Deficiency in the ubiquitin conjugating enzyme UBE2A in alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel). 7, 116 (2016).
Bosco, P., Spada, R., Caniglia, S., Salluzzo, M. G. & Salemi, M. Cerebellar degeneration-related autoantigen 1 (CDR1) gene expression in Alzheimer's disease. Neurol. Sci. 35, 1613–1614 (2014).
Morais, V. A. et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344, 203–207 (2014).
Scheele, C. et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8, 74 (2007).
Muddashetty, R. S. et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321, 433–445 (2002).
Mus, E., Hof, P. R. & Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl Acad. Sci. USA 104, 10679–10684 (2007).
Liu, Y. et al. Association between ubiquitin carboxy-terminal hydrolase-L1 S18Y variant and risk of Parkinson's disease: the impact of ethnicity and onset age. Neurol. Sci. 36, 179–188 (2015).
Carrieri, C. et al. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells' differentiation in vitro and in neurochemical models of Parkinson's disease. Front. Cell. Neurosci. 9, 114 (2015).
Ambrosi, G. et al. Bioenergetic and proteolytic defects in fibroblasts from patients with sporadic Parkinson's disease. Biochim. Biophys. Acta - Mol. Basis Dis. 1842, 1385–1394 (2014).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A. ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146, 353–358 (2011).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283 (2016).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). This is one of the first reports on the role of circRNAs as miRNA sponges.
Zhang, K. et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015).
Tollervey, J. R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011).
Adriaens, C. et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 22, 861–868 (2016).
Yamanaka, Y. et al. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein. HMGB2. Cell Rep. 11, 967–976 (2015).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). This is one of the first papers on the role of circRNAs as miRNA sponges.
Shi, Z. et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-κB-dependent manner. FEBS J. 284, 1096–1109 (2017).
Junn, E. et al. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl Acad. Sci. USA 106, 13052–13057 (2009).
Tan, J. Y. et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat. Struct. Mol. Biol. 21, 955–961 (2014).
Kumar, V. et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 9, e1003201 (2013).
Shi, J. et al. A 3′-UTR polymorphism in the oxidized LDL receptor 1 gene increases Abeta40 load as cerebral amyloid angiopathy in Alzheimer's disease. Acta Neuropathol. 111, 15–20 (2006).
Wang, G. et al. Variation in the miRNA-433 binding site of FGF20 confers risk for parkinson disease by overexpression of α-synuclein. Am. J. Hum. Genet. 82, 283–289 (2008).
Rademakers, R. et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum. Mol. Genet. 17, 3631–3642 (2008).
Rollinson, S. et al. No association of PGRN 3′UTR rs5848 in frontotemporal lobar degeneration. Neurobiol. Aging 32, 754–755 (2011).
Simón-Sánchez, J. et al. Variation at GRN 3′-UTR rs5848 is not associated with a risk of frontotemporal lobar degeneration in Dutch population. PLoS ONE 4, e7494 (2009).
Hsiung, G.-Y. R., Fok, A., Feldman, H. H., Rademakers, R. & Mackenzie, I. R. A. rs5848 polymorphism and serum progranulin level. J. Neurol. Sci. 300, 28–32 (2011).
Nelson, P. T. et al. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J. Neuropathol. Exp. Neurol. 74, 75–84 (2015).
Zhang, B., Wang, A., Xia, C., Lin, Q. & Chen, C. A single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer's disease and is associated with the pathogenesis of Alzheimer's disease. Mol. Med. Rep. 12, 4037–4042 (2015).
Vermunt, M. W. et al. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 9, 767–779 (2014).
Soldner, F. et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016).
Bhartiya, D. & Scaria, V. Genomic variations in non-coding RNAs: structure, function and regulation. Genomics 107, 59–68 (2016).
Qureshi, I. A. & Mehler, M. F. Epigenetic mechanisms governing the process of neurodegeneration. Mol. Aspects Med. 34, 875–882 (2013).
Chevillet, J. R., Lee, I., Briggs, H. A., He, Y. & Wang, K. Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19, 6080–6105 (2014).
Sala Frigerio, C. et al. Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81, 2103–2106 (2013).
Dong, H. et al. Serum microRNA profiles serve as novel biomarkers for the diagnosis of alzheimer's disease. Dis. Markers 2015, 625659 (2015).
Cheng, L. et al. Prognostic serum miRNA biomarkers associated with Alzheimer's disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1–9 (2014).
Alvarez-Mora, M. I. et al. MicroRNA expression profiling in blood from fragile X-associated tremor/ataxia syndrome patients. Genes Brain Behav. 12, 595–603 (2013).
Gandhi, R. et al. Circulating MicroRNAs as biomarkers for disease staging in multiple sclerosis. Ann. Neurol. 73, 729–740 (2013).
Grasso, M., Piscopo, P., Confaloni, A. & Denti, M. A. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules 19, 6891–6910 (2014).
Denk, J. et al. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer's disease. PLoS ONE 10, 1–18 (2015).
Moon, J. et al. Early diagnosis of Alzheimer's disease from elevated olfactory mucosal miR-206 level. Sci. Rep. 6, 20364 (2016).
Su, Z. et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83, 1043–1050 (2014).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Lau, P., Sala Frigerio, C. & De Strooper, B. Variance in the identification of microRNAs deregulated in Alzheimer's disease and possible role of lincRNAs in the pathology: The need of larger datasets. Ageing Res. Rev. 17, 43–53 (2014).
Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 12, 433–446 (2013). This is one of the first reports on therapeutic targeting of lncRNAs.
DeVos, S. L. & Miller, T. M. Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics 10, 486–497 (2013).
Donnelly, C. J. et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013). This is one of the first papers to propose RNA targeting as a therapeutic approach in ALS and FTD.
Sareen, D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 5, 208ra149 (2013).
Jiang, J. et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90, 535–550 (2016).
Kramer, N. J. et al. Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science 353, 708–712 (2016).
Modarresi, F. et al. Knockdown of BACE1-AS nonprotein-coding transcript modulates ß-amyloid-related hippocampal neurogenesis. Int. J. Alzheimers. Dis. 2011, 929042 (2011).
Armakola, M. et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 44, 1302–1309 (2012).
DeVos, S. L. & Miller, T. M. Direct intraventricular delivery of drugs to the rodent central nervous system. J. Vis. Exp. http://dx.doi.org/10.3791/50326 (2013).
Schoch, K. M. et al. Increased 4R-Tau induces pathological changes in a human-tau mouse model. Neuron 90, 941–947 (2016).
DeVos, S. L. et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 9, eaag0481 (2017).
Smith, R. A. et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 116, 2290–2296 (2006).
Hua, Y. et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010).
Wahlgren, J. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 40, e130 (2012).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Munoz, J. L. et al. Delivery of functional anti-miR-9 by mesenchymal Stem cell–derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2, e126 (2013).
Alvarez-Erviti, L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345 (2011).
Matsui, M. & Corey, D. R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16, 167–179 (2016).
Miller, T. M. et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 12, 435–442 (2013). This is the first report on a clinical trial using ASOs in neurodegeneration.
Finkel, R. S. et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388, 3017–3026 (2016). This paper reports the very recent results from a successful clinical trial using ASOs in infants with SMA.
Lardenoije, R. et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 131, 21–64 (2015).
Zhang, R., Deng, P., Jacobson, D. & Li, J. B. Evolutionary analysis reveals regulatory and functional landscape of coding and non-coding RNA editing. PLoS Genet. 13, e1006563 (2017).
Guffanti, A., Simchovitz, A. & Soreq, H. Emerging bioinformatics approaches for analysis of NGS-derived coding and non-coding RNAs in neurodegenerative diseases. Front. Cell. Neurosci. 8, 89 (2014).
da Sacco, L., Baldassarre, A. & Masotti, A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int. J. Mol. Sci. 13, 97–114 (2012).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). This is one of the first systematic studies of spatial transcriptomics technology.
Leone, S. & Santoro, R. Challenges in the analysis of long noncoding RNA functionality. FEBS Lett. 590, 2342–2353 (2016).
Goyal, A. et al. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45, e12 (2016).
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015).
Bergmann, J. H. & Spector, D. L. Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18 (2014).
Brown, C. J. et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 (1992).
Ip, J. Y. & Nakagawa, S. Long non-coding RNAs in nuclear bodies. Dev. Growth Differ. 54, 44–54 (2012).
Chu, C., Spitale, R. C. & Chang, H. Y. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 29–35 (2015).
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016).
Engreitz, J. M., Ollikainen, N. & Guttman, M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell. Biol. 17, 756–770 (2016).
Yang, Y., Wen, L. & Zhu, H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 5, 59 (2015).
Ferrè, F., Colantoni, A. & Helmer-Citterich, M. Revealing protein–lncRNA interaction. Brief. Bioinform. 17, 106–116 (2016).
McHugh, C. A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015).
Chu, C. et al. Systematic discovery of xist RNA binding proteins. Cell 161, 404–416 (2015).
Quinn, J. J. et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 32, 933–940 (2014).
Minajigi, A. et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 (2015).
Bartonicek, N., Maag, J. L. V. & Dinger, M. E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer 15, 43 (2016).
Buenrostro, J. D. et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat. Biotechnol. 32, 562–568 (2014).
Loya, C. M., Lu, C. S., Van Vactor, D. & Fulga, T. A. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897–903 (2009).
Meng, L. et al. Small RNA zippers lock miRNA molecules and block miRNA function in mammalian cells. Nat. Commun. 8, 13964 (2017).
Bassett, A. R. et al. Understanding functional miRNA–target interactions in vivo by site-specific genome engineering. Nat. Commun. 5, 4640 (2014).
Klann, T. S. et al. CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561–568 (2017).
Dinger, M. E., Pang, K. C., Mercer, T. R. & Mattick, J. S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4, e1000176 (2008).
Filippenkov, I. B., Kalinichenko, E. O., Limborska, S. A. & Dergunova, L. V. Circular RNAs-one of the enigmas of the brain. Neurogenetics 18, 1–6 (2016).
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733 (2012).
Ruiz-Orera, J. et al. Long non-coding RNAs as a source of new peptides. eLife 3, e03523 (2014).
van Heesch, S. et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15, R6 (2014).
Niazi, F. & Valadkhan, S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. Rna 18, 825–843 (2012).
Guttman, M., Russell, P., Ingolia, N. T., Weissman, J. S. & Lander, E. S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251 (2013).
Lauressergues, D. et al. Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93 (2015). This paper reports the protein-coding potential of miRNA primary transcripts.
Gendron, T. F. et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). This is one of the first reports of antisense RAN translation from the C9ORF72 locus.
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Legnini, I. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9 (2017).
Pamudurti, N. R. et al. Translation of circRNAs. Mol. Cell 66, 9–21.e7 (2017).
Zhao, Y. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, D203–D208 (2016).
Qureshi, I. A. & Mehler, M. F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 13, 528–541 (2012).
ENCODE Project et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Rands, C. M., Meader, S., Ponting, C. P. & Lunter, G. 8.2% of the human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 10, e1004525 (2014).
Graur, D. et al. On the immortality of television sets: 'function' in the human genome according to the evolution-free gospel of encode. Genome Biol. Evol. 5, 578–590 (2013).
Hon, C.-C. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017). This is the most recent systematic documentation of human lncRNAs.
Taft, R. J., Pheasant, M. & Mattick, J. S. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29, 288–299 (2007).
Liu, G., Mattick, J. S. & Taft, R. J. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 12, 2061–2072 (2013).
Mattick, J. S. RNA regulation: a new genetics? Nat. Rev. Genet. 5, 316–323 (2004).
Bentwich, I. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37, 766–770 (2005).
Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011).
Qureshi, I. A. & Mehler, M. F. An evolving view of epigenetic complexity in the brain. Phil. Trans. R. Soc. B Biol. Sci. 369, 1–8 (2014).
Prabhakar, S., Noonan, J. P., Pääbo, S. & Rubin, E. M. Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (2006).
Hu, H. Y. et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 7, 13–15 (2011).
Clark, B. S. & Blackshaw, S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front. Genet. 5, 1–19 (2014).
Ataman, B. et al. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247 (2016).
Kwan, K. Y. et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911 (2012).
Johnson, M., Kawasawa, Y. & Mason, C. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009).
Pollard, K. S. et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172 (2006). This is the first report on a human-specific and brain-specific ncRNA.
Somel, M. et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 9, e1001214 (2011).
Somel, M., Liu, X. & Khaitovich, P. Human brain evolution: transcripts, metabolites and their regulators. Nat. Rev. Neurosci. 14, 112–127 (2013).
Wood, S. H., Craig, T., Li, Y., Merry, B. & De Magalhães, J. P. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age 35, 763–776 (2013).
Persengiev, S., Kondova, I., Otting, N., Koeppen, A. H. & Bontrop, R. E. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 32, 2316.e17 (2011).
Lipovich, L. et al. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb. Cortex 24, 1451–1459 (2013).
Finch, C. E. & Austad, S. N. Primate aging in the mammalian scheme: the puzzle of extreme variation in brain aging. Age 34, 1075–1091 (2012).
Coulson, E. J., Paliga, K., Beyreuther, K. & Masters, C. L. What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem. Int. 36, 175–184 (2000).
Holzer, M., Craxton, M., Jakes, R., Arendt, T. & Goedert, M. Tau gene (MAPT) sequence variation among primates. Gene 341, 313–322 (2004).
Siddiqui, I. J., Pervaiz, N. & Abbasi, A. A. The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci. Rep. 6, 24475 (2016).
Ruzo, A. et al. Discovery of novel isoforms of Huntingtin reveals a new hominid-specific exon. PLoS ONE 10, 1–13 (2015).
Johnson, R. et al. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol. Genom. 41, 269–274 (2010).
Espuny-Camacho, I. et al. Hallmarks of alzheimer's disease in stem-cell-derived human neurons transplanted into mouse brain. Neuron 93, 1066–1081.e8 (2017). This is the first study using an induced pluripotent stem cell-based in vivo humanized chimeric human-mouse AD model to demonstrate the human-specific requirements for neurodegeneration.
Hagerman, P. J. & Hagerman, R. J. Fragile X-associated tremor/ataxia syndrome. Ann. NY Acad. Sci. 1338, 58–70 (2015).
Femminella, G. D., Ferrara, N. & Rengo, G. The emerging role of microRNAs in Alzheimer's disease. Front. Physiol. 6, 1–5 (2015).
Lee, Y. et al. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nature Neurosci. 11, 1137–1139 (2008).
da Silva, F. et al. microRNAs involved in Parkinson's disease: a systematic review. Mol. Med. Rep. http://dx.doi.org/10.3892/mmr.2016.5759 (2016).
Tan, H., Poidevin, M., Li, H., Chen, D. & Jin, P. MicroRNA-277 modulates the neurodegeneration caused by fragile X premutation rCGG repeats. PLoS Genet. 8, e1002681 (2012).
Zongaro, S. et al. The 3′ UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum. Mol. Genet. 22, 1971–1982 (2013).
Gaughwin, P. M. et al. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum. Mol. Genet. 20, 2225–2237 (2011).
Johnson, R. et al. Regulation of neural macroRNAs by the transcriptional repressor REST. Rna 15, 85–96 (2009).
Hoss, A G. et al. miR-10b-5p expression in Huntington's disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genomics 8, 10 (2015).
Packer, A. N., Xing, Y., Harper, S. Q., Jones, L. & Davidson, B. L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 28, 14341–14346 (2008).
Gascon, E. & Gao, F.-B. The emerging roles of MicroRNAs in the pathogenesis of frontotemporal dementia–amyotrophic lateral sclerosis (FTD-ALS) spectrum disorders. J. Neurogenet. 28, 30–40 (2014).
Acknowledgements
E.S. receives funding from the Fonds voor Wetenschappelijk Onderzoek (FWO) and the Alzheimer's Association. B.D.S. was supported by a European Research Council (ERC) grant for his miRNA work and is supported by the FWO, KU Leuven, VIB, and a Methusalem grant from KU Leuven and the Flemish Government. He is further supported by the Opening the Future campaign of the Leuven Universiteit Fonds (LUF). The authors are grateful to C. Sala Frigerio, E. Leucci, A. Sierksma, R. Guerreiro and J. Bras for reading the manuscript and providing critical feedback.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
ncRNAs in neurodegeneration. (PDF 1841 kb)
Glossary
- Long non-coding RNAs
-
(lncRNAs). Non-protein-coding transcripts that are longer than 200–400 nucleotides and include multiple diverse RNA species.
- Circular RNAs
-
(circRNAs). Covalently closed, single-stranded transcripts that are produced by the back-splicing of exons in precursor mRNAs.
- microRNAs
-
(miRNAs). Small (20–25 nucleotides) non-protein-coding regulatory RNA molecules that are involved in post-transcriptional regulation.
- Endogenous small-interfering RNAs
-
(endo-siRNAs). Small (21–26 nucleotides) non-protein-coding regulatory RNAs that are produced from endogenous double-stranded RNA precursors and are involved in post-transcriptional silencing.
- Small nucleolar-derived RNAs
-
Small (17–30 nucleotides) non-protein-coding regulatory RNAs that are derived from the processing of small nucleolar RNAs and are implicated in gene silencing.
- PIWI-interacting RNAs
-
(piRNAs). Small (26–33 nucleotides) non-protein-coding regulatory RNAs involved in epigenetic and post-transcriptional gene silencing via interaction with PIWI proteins.
- Long natural antisense transcripts
-
(NATs). Long (200–400 nucleotides) RNA molecules that are transcribed from the opposite DNA strand; they partially overlap with the sense transcript and often regulate its transcription, splicing or stability.
- Enhancer non-coding RNAs
-
(eRNAs). Non-protein-coding RNAs that are transcribed from enhancer DNA loci and are implicated in the regulation of gene transcription.
- Convergent transcription
-
Simultaneous transcription proceeding in the sense and antisense orientation from two closely positioned promoters, with the RNA polymerases heading towards each other.
- Frontotemporal lobar degeneration with TAR DNA-binding protein 43 (TDP43) proteinopathy
-
(FTLD-TDP). Frontotemporal lobar degeneration with tau-negative, ubiquitin-positive inclusions that contain TDP43.
- Seed sequence
-
Nucleotide sequences (2–7 nucleotides) in the 5′-end of the microRNA (miRNA) sequence that are crucial for recognizing and binding to complementary sites on target mRNA 3′-untranslated regions (UTRs).
Rights and permissions
About this article
Cite this article
Salta, E., De Strooper, B. Noncoding RNAs in neurodegeneration. Nat Rev Neurosci 18, 627–640 (2017). https://doi.org/10.1038/nrn.2017.90
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn.2017.90
This article is cited by
-
Neuroinflammation driven by human immunodeficiency virus-1 (HIV-1) directs the expression of long noncoding RNA RP11-677M14.2 resulting in dysregulation of neurogranin in vivo and in vitro
Journal of Neuroinflammation (2024)
-
Eip74EF is a dominant modifier for ALS-FTD-linked VCPR152H phenotypes in the Drosophila eye model
BMC Research Notes (2023)
-
Epigenetic regulation in major depression and other stress-related disorders: molecular mechanisms, clinical relevance and therapeutic potential
Signal Transduction and Targeted Therapy (2023)
-
Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets
Functional & Integrative Genomics (2023)
-
Discerning the Prospects of miRNAs as a Multi-Target Therapeutic and Diagnostic for Alzheimer’s Disease
Molecular Neurobiology (2023)