Abstract
According to current paradigms, various risk factors, such as genetic mutations, oxidative stress, neural network dysfunction, and abnormal protein degradation, contribute to the progression of brain disorders. Through the cooperation of gene transcripts in biological processes, the study of noncoding RNAs can lead to insights into the cause and treatment of brain disorders. Recently, long noncoding RNAs (lncRNAs) which are longer than 200 nucleotides in length have been suggested as key factors in various brain disorders. Accumulating evidence suggests the potential of lncRNAs as diagnostic or prognostic biomarkers and therapeutic targets. High-throughput screening-based sequencing has been instrumental in identification of lncRNAs that demand new approaches to understanding the progression of brain disorders. In this review, we discuss the recent progress in the study of lncRNAs, and addresses the pathogenesis of brain disorders that involve lncRNAs and describes the associations of lncRNAs with neurodegenerative disorders such as Alzheimer disease (AD), Parkinson disease (PD), and neurodevelopmental disorders. We also discuss potential targets of lncRNAs and their promise as novel therapeutics and biomarkers in brain disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci USA. 2013;110:5294–300.
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–55.
Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44.
Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5.
Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol Cell. 2018;72:970–84.e77.
Sati S, Ghosh S, Jain V, Scaria V, Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012;40:10018–31.
Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–8.
Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19:535–48.
Kim TK, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol. 2015;7:a018622.
Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat Neurosci. 2016;19:75–83.
Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42.
Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162:948–59.
Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89:283–308.
Salta E, De Strooper B. Noncoding RNAs in neurodegeneration. Nat Rev Neurosci. 2017;18:627–40.
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26.
Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra140.
Velmeshev D, Magistri M, Faghihi MA. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol Autism. 2013;4:32.
Liu Y, Chen YY, Liu H, Yao CJ, Zhu XX, Chen DJ, et al. Association between ubiquitin carboxy-terminal hydrolase-L1 S18Y variant and risk of Parkinson’s disease: the impact of ethnicity and onset age. Neurol Sci. 2015;36:179–88.
Carrieri C, Forrest AR, Santoro C, Persichetti F, Carninci P, Zucchelli S, et al. Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci. 2015;9:114.
Tromp D, Dufour A, Lithfous S, Pebayle T, Despres O. Episodic memory in normal aging and Alzheimer disease: Insights from imaging and behavioral studies. Ageing Res Rev. 2015;24:232–62.
Galvao F Jr, Grokoski KC, da Silva BB, Lamers ML, Siqueira IR. The amyloid precursor protein (APP) processing as a biological link between Alzheimer’s disease and cancer. Ageing Res Rev. 2019;49:83–91.
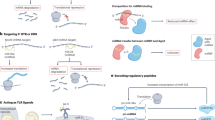
Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30.
Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56.
Zeng T, Ni H, Yu Y, Zhang M, Wu M, Wang Q, et al. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J Chem Neuroanat. 2019;98:87–96.
Das B, Yan R. Role of BACE1 in Alzheimer’s synaptic function. Transl Neurodegener. 2017;6:23.
Wang P, Zheng X, Guo Q, Yang P, Pang X, Qian K, et al. Systemic delivery of BACE1 siRNA through neuron-targeted nanocomplexes for treatment of Alzheimer’s disease. J Control Release. 2018;279:220–33.
Sosinska P, Mikula-Pietrasik J, Ksiazek K. The double-edged sword of long non-coding RNA: the role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer. Mutat Res Rev Mutat Res. 2015;766:58–67.
Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–51.
Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, et al. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262.
Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16:109.
Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci USA. 2007;104:10679–84.
Li H, Zheng L, Jiang A, Mo Y, Gong Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport. 2018;29:1061–7.
Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem Res. 1992;17:591–7.
Fatemi SH, Folsom TD, Reutiman TJ, Thuras PD. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum. 2009;8:64–9.
Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308–17.
Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, et al. An intronic ncRNA-dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis Model Mech. 2013;6:424–33.
K TC, Lunetta KL, Baldwin CT, McKee AC, Guo J, Cupples LA, et al. Association of distinct variants in SORL1 with cerebrovascular and neurodegenerative changes related to Alzheimer disease. Arch Neurol. 2008;65:1640–8.
Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–77.
Deng YY, Xiao L, Li W, Tian M, Feng XL, Feng H, et al. Plasma long noncoding RNA 51A as a stable biomarker of Alzheimer’s disease. Int J Clin Exp Pathol. 2017;10:4694–9.
Song M, Martinowich K, Lee FS. BDNF at the synapse: why location matters. Mol Psychiatry. 2017;22:1370–5.
Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–22.
Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–9.
Guo CC, Jiao CH, Gao ZM. Silencing of LncRNA BDNF-AS attenuates Abeta25-35-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol Res. 2018;40:795–804.
Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603.
Parenti R, Paratore S, Torrisi A, Cavallaro S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur J Neurosci. 2007;26:2444–57.
Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. 2010;24:4033–46.
Massone S, Ciarlo E, Vella S, Nizzari M, Florio T, Russo C, et al. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid beta secretion. Biochim Biophys Acta. 2012;1823:1170–7.
Kinney JW, Bemiller SM, Murtishaw AS, Leisgang AM, Salazar AM, Lamb BT. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 2018;4:575–90.
Clarkson ED, Zawada WM, Freed CR. GDNF improves survival and reduces apoptosis in human embryonic dopaminergic neurons in vitro. Cell Tissue Res. 1997;289:207–10.
Ayanlaja AA, Zhang B, Ji G, Gao Y, Wang J, Kanwore K, et al. The reversible effects of glial cell line-derived neurotrophic factor (GDNF) in the human brain. Semin Cancer Biol. 2018;53:212–22.
Sherer TB, Fiske BK, Svendsen CN, Lang AE, Langston JW. Crossroads in GDNF therapy for Parkinson’s disease. Mov Disord. 2006;21:136–41.
Gill SS, Patel NK, Hotton GR, O’Sullivan K, McCarter R, Bunnage M, et al. Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat Med. 2003;9:589–95.
Straten G, Eschweiler GW, Maetzler W, Laske C, Leyhe T. Glial cell-line derived neurotrophic factor (GDNF) concentrations in cerebrospinal fluid and serum of patients with early Alzheimer’s disease and normal controls. J Alzheimers Dis. 2009;18:331–7.
Airavaara M, Pletnikova O, Doyle ME, Zhang YE, Troncoso JC, Liu QR. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J Biol Chem. 2011;286:45093–102.
Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Prim. 2017;3:17013.
DeMaagd G, Philip A. Parkinson’s disease and its management: part 1: disease entity, risk factors, pathophysiology, clinical presentation, and diagnosis. P T. 2015;40:504–32.
Chaudhuri KR, Healy DG, Schapira AH. National Institute for Clinical E. Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol. 2006;5:235–45.
Stefanis L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a009399.
Elkouris M, Kouroupi G, Vourvoukelis A, Papagiannakis N, Kaltezioti V, Matsas R, et al. Long non-coding RNAs associated with neurodegeneration-linked genes are reduced in parkinson’s disease patients. Front Cell Neurosci. 2019;13:58.
Lim KH, Joo JY, Baek KH. The potential roles of deubiquitinating enzymes in brain diseases. Ageing Res Rev. 2020;61:101088.
Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, et al. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci USA. 2013;110:3489–94.
Liu H, Povysheva N, Rose ME, Mi Z, Banton JS, Li W, et al. Role of UCHL1 in axonal injury and functional recovery after cerebral ischemia. Proc Natl Acad Sci USA. 2019;116:4643–50.
Healy DG, Abou-Sleiman PM, Casas JP, Ahmadi KR, Lynch T, Gandhi S, et al. UCHL-1 is not a Parkinson’s disease susceptibility gene. Ann Neurol. 2006;59:627–33.
Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–18.
Maraganore DM, Lesnick TG, Elbaz A, Chartier-Harlin MC, Gasser T, Kruger R, et al. UCHL1 is a Parkinson’s disease susceptibility gene. Ann Neurol. 2004;55:512–21.
Riva P, Ratti A, Venturin M. The Long Non-Coding RNAs in Neurodegenerative Diseases: Novel Mechanisms of Pathogenesis. Curr Alzheimer Res. 2016;13:1219–31.
Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7.
Deas E, Plun-Favreau H, Wood NW. PINK1 function in health and disease. EMBO Mol Med. 2009;1:152–65.
Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–73.
Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60.
Simchovitz A, Hanan M, Niederhoffer N, Madrer N, Yayon N, Bennett ER, et al. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. 2019;33:11223–34.
Scheele C, Petrovic N, Faghihi MA, Lassmann T, Fredriksson K, Rooyackers O, et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genom. 2007;8:74.
Boros FA, Maszlag-Torok R, Vecsei L, Klivenyi P. Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res. 2020;1730:146672.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Lang Y, Li Y, Yu H, Lin L, Chen X, Wang S, et al. HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson’s disease by elevating NPTX2 via miR-221-3p binding. Aging. 2020;12:7660–78.
Lin Q, Hou S, Dai Y, Jiang N, Lin Y. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem. 2019;400:1217–28.
Shirvani Farsani Z, Zahirodin A, Ghaderian SMH, Shams J, Naghavi Gargari B. The role of long non-coding RNA MALAT1 in patients with bipolar disorder. Metab Brain Dis. 2020;35:1077–83.
Kouroupi G, Taoufik E, Vlachos IS, Tsioras K, Antoniou N, Papastefanaki F, et al. Defective synaptic connectivity and axonal neuropathology in a human iPSC-based model of familial Parkinson’s disease. Proc Natl Acad Sci USA. 2017;114:E3679–88.
Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14.
Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA. Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol Neurobiol. 2017;54:2869–77.
Liu W, Zhang Q, Zhang J, Pan W, Zhao J, Xu Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017;7:19.
Satoh J, Tabunoki H, Arima K. Molecular network analysis suggests aberrant CREB-mediated gene regulation in the Alzheimer disease hippocampus. Dis Markers. 2009;27:239–52.
Yao J, Wang XQ, Li YJ, Shan K, Yang H, Wang YN, et al. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med. 2016;8:346–62.
Tobin JE, Latourelle JC, Lew MF, Klein C, Suchowersky O, Shill HA, et al. Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology. 2008;71:28–34.
Coupland KG, Kim WS, Halliday GM, Hallupp M, Dobson-Stone C, Kwok JB. Role of the long non-coding RNA MAPT-AS1 in regulation of microtubule associated protein tau (MAPT) expression in Parkinson’s disease. PLoS ONE. 2016;11:e0157924.
Das G, Misra AK, Das SK, Ray K, Ray J. Microtubule-associated protein tau (MAPT) influences the risk of Parkinson’s disease among Indians. Neurosci Lett. 2009;460:16–20.
Ni Y, Huang H, Chen Y, Cao M, Zhou H, Zhang Y. Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell Mol Neurobiol. 2017;37:329–38.
Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 Suppl:S10–17.
De Souza RA, Leavitt BR. Neurobiology of Huntington’s disease. Curr Top Behav Neurosci. 2015;22:81–100.
Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, et al. Huntington disease. Nat Rev Dis Prim. 2015;1:15005.
Wild EJ, Tabrizi SJ. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16:837–47.
Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet. 2011;20:3467–77.
Hwang JY, Zukin RS. REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol. 2018;48:193–200.
Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, et al. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genom. 2010;41:269–74.
Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–5.
Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36:1643–51.
Li Q, Zhang J, Su DM, Guan LN, Mu WH, Yu M, et al. lncRNA TUG1 modulates proliferation, apoptosis, invasion, and angiogenesis via targeting miR-29b in trophoblast cells. Hum Genom. 2019;13:50.
Chen C, Cheng G, Yang X, Li C, Shi R, Zhao N. Tanshinol suppresses endothelial cells apoptosis in mice with atherosclerosis via lncRNA TUG1 up-regulating the expression of miR-26a. Am J Transl Res. 2016;8:2981–91.
Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46:245–54.
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72.
Sunwoo JS, Lee ST, Im W, Lee M, Byun JI, Jung KH, et al. Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol Neurobiol. 2017;54:1577–86.
Cheng C, Spengler RM, Keiser MS, Monteys AM, Rieders JM, Ramachandran S, et al. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum Mol Genet. 2018;27:4303–14.
Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–47.
Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an oncogenic long non-coding RNA in different cancers. Pathol Oncol Res. 2019;25:859–74.
Chanda K, Das S, Chakraborty J, Bucha S, Maitra A, Chatterjee R, et al. Altered levels of long NcRNAs Meg3 and Neat1 in cell and animal models of Huntington’s disease. RNA Biol. 2018;15:1348–63.
Brochier C, Gaillard MC, Diguet E, Caudy N, Dossat C, Segurens B, et al. Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in human and affected in disease models. Physiol Genom. 2008;33:170–9.
Lord CC, Thomas G, Brown JM. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim Biophys Acta. 2013;1831:792–802.
Zhuang X, Tong H, Ding Y, Wu L, Cai J, Si Y, et al. Long noncoding RNA ABHD11-AS1 functions as a competing endogenous RNA to regulate papillary thyroid cancer progression by miR-199a-5p/SLC1A5 axis. Cell Death Dis. 2019;10:620.
Park H, Miyazaki H, Yamanaka T, Nukina N. Non-coding RNA Neat1 and Abhd11os expressions are dysregulated in medium spiny neurons of Huntington disease model mice. Neurosci Res. 2019;147:58–63.
Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA. 1997;278:1349–56.
Rojas C, Barnaeva E, Thomas AG, Hu X, Southall N, Marugan J, et al. DPTIP, a newly identified potent brain penetrant neutral sphingomyelinase 2 inhibitor, regulates astrocyte-peripheral immune communication following brain inflammation. Sci Rep. 2018;8:17715.
Adams JS, Adams PE, Nguyen D, Brunberg JA, Tassone F, Zhang W, et al. Volumetric brain changes in females with fragile X-associated tremor/ataxia syndrome (FXTAS). Neurology. 2007;69:851–9.
Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19.
Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–87.
Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet. 2014;133:59–67.
Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–96.
Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50.
d’Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, et al. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. 2017;93:66–79.
Corral-Juan M, Serrano-Munuera C, Rabano A, Cota-Gonzalez D, Segarra-Roca A, Ispierto L, et al. Clinical, genetic and neuropathological characterization of spinocerebellar ataxia type 37. Brain. 2018;141:1981–97.
Klockgether T, Mariotti C, Paulson HL. Spinocerebellar ataxia. Nat Rev Dis Prim. 2019;5:24.
Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544:362–6.
Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, Curtis HJ, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21:955–61.
Evert BO, Nalavade R, Jungverdorben J, Matthes F, Weber S, Rajput A, et al. Upregulation of miR-370 and miR-543 is associated with reduced expression of heat shock protein 40 in spinocerebellar ataxia type 3. PLoS ONE. 2018;13:e0201794.
Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–20.
Aleman TS, Cideciyan AV, Volpe NJ, Stevanin G, Brice A, Jacobson SG. Spinocerebellar ataxia type 7 (SCA7) shows a cone-rod dystrophy phenotype. Exp Eye Res. 2002;74:737–45.
Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–84.
Sparber P, Filatova A, Khantemirova M, Skoblov M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med Genom. 2019;12:42.
Ayhan F, Perez BA, Shorrock HK, Zu T, Banez-Coronel M, Reid T, et al. SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by eIF3F. EMBO J. 2018;37:e99023.
Sawada J, Katayama T, Tokashiki T, Kikuchi S, Kano K, Takahashi K, et al. The first case of spinocerebellar ataxia type 8 in monozygotic twins. Intern Med. 2020;59:277–83.
Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600.
Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–69.
Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Prim. 2017;3:17071.
van Es MA, Hardiman O, Chio A, Al-Chalabi A, Pasterkamp RJ, Veldink JH, et al. Amyotrophic lateral sclerosis. Lancet. 2017;390:2084–98.
Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104.
Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–44.
Nishimoto Y, Nakagawa S, Hirose T, Okano HJ, Takao M, Shibata S, et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain. 2013;6:31.
Corrado L, Mazzini L, Oggioni GD, Luciano B, Godi M, Brusco A, et al. ATXN-2 CAG repeat expansions are interrupted in ALS patients. Hum Genet. 2011;130:575–80.
Toyoshima Y, Tanaka H, Shimohata M, Kimura K, Morita T, Kakita A, et al. Spinocerebellar ataxia type 2 (SCA2) is associated with TDP-43 pathology. Acta Neuropathol. 2011;122:375–8.
Li PP, Sun X, Xia G, Arbez N, Paul S, Zhu S, et al. ATXN2-AS, a gene antisense to ATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann Neurol. 2016;80:600–15.
Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–58.
Amick J, Ferguson SM. C9orf72: At the intersection of lysosome cell biology and neurodegenerative disease. Traffic. 2017;18:267–76.
Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38.
Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530–9.
Gagliardi S, Pandini C, Garofalo M, Bordoni M, Pansarasa O, Cereda C. Long non coding RNAs and ALS: Still much to do. Noncoding RNA Res. 2018;3:226–31.
Riboldi G, Zanetta C, Ranieri M, Nizzardo M, Simone C, Magri F, et al. Antisense oligonucleotide therapy for the treatment of C9ORF72 ALS/FTD diseases. Mol Neurobiol. 2014;50:721–32.
Smigielski L, Jagannath V, Rossler W, Walitza S, Grunblatt E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol Psychiatry. 2020;25:1718–48.
Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97.
Kesby JP, Eyles DW, McGrath JJ, Scott JG. Dopamine, psychosis and schizophrenia: the widening gap between basic and clinical neuroscience. Transl Psychiatry. 2018;8:30.
Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA. 2009;106:15873–8.
Chung DW, Volk DW, Arion D, Zhang Y, Sampson AR, Lewis DA. Dysregulated ErbB4 Splicing in Schizophrenia: Selective Effects on Parvalbumin Expression. Am J Psychiatry. 2016;173:60–8.
Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19:486–94.
Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–90.
Vieta E, Berk M, Schulze TG, Carvalho AF, Suppes T, Calabrese JR, et al. Bipolar disorders. Nat Rev Dis Prim. 2018;4:18008.
Phillips ML, Kupfer DJ. Bipolar disorder diagnosis: challenges and future directions. Lancet. 2013;381:1663–71.
Harrison PJ, Geddes JR, Tunbridge EM. The emerging neurobiology of bipolar disorder. Trends Neurosci. 2018;41:18–30.
Prisciandaro JJ, Tolliver BK, Prescot AP, Brenner HM, Renshaw PF, Brown TR, et al. Unique prefrontal GABA and glutamate disturbances in co-occurring bipolar disorder and alcohol dependence. Transl Psychiatry. 2017;7:e1163.
Ryan NM, Lihm J, Kramer M, McCarthy S, Morris SW, Arnau-Soler A, et al. DNA sequence-level analyses reveal potential phenotypic modifiers in a large family with psychiatric disorders. Mol Psychiatry. 2018;23:2254–65.
Millar JK, James R, Brandon NJ, Thomson PA. DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric illness. Ann Med. 2004;36:367–78.
Tropea D, Hardingham N, Millar K, Fox K. Mechanisms underlying the role of DISC1 in synaptic plasticity. J Physiol. 2018;596:2747–71.
Naghavi-Gargari B, Zahirodin A, Ghaderian SMH, Shirvani-Farsani Z. Significant increasing of DISC2 long non-coding RNA expression as a potential biomarker in bipolar disorder. Neurosci Lett. 2019;696:206–11.
Luykx JJ, Giuliani F, Giuliani G, Veldink J. Coding and non-coding RNA abnormalities in bipolar disorder. Genes 2019;10:946–60.
Garcia JC, Bustos RH. The genetic diagnosis of neurodegenerative diseases and therapeutic perspectives. Brain Sci. 2018;8:222–40.
Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–77.
Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-targeted therapeutics. Cell Metab. 2018;27:714–39.
Wurster CD, Ludolph AC. Antisense oligonucleotides in neurological disorders. Ther Adv Neurol Disord. 2018;11:1756286418776932.
Buiting K, Williams C, Horsthemke B. Angelman syndrome—insights into a rare neurogenetic disorder. Nat Rev Neurol. 2016;12:584–93.
Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–12.
Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11:63.
Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9.
Zhou Y, Shan T, Ding W, Hua Z, Shen Y, Lu Z, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo-YAP signaling. J Cell Physiol. 2018;233:5805–14.
Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018;32:1948–57.
Raymond GJ, Zhao HT, Race B, Raymond LD, Williams K, Swayze EE, et al. Antisense oligonucleotides extend survival of prion-infected mice. JCI Insight. 2019;5:e131175.
Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377:1723–32.
Talbot K, Tizzano EF. The clinical landscape for SMA in a new therapeutic era. Gene Ther. 2017;24:529–33.
Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, et al. Targeting Huntingtin expression in patients with Huntington’s disease. N Engl J Med. 2019;380:2307–16.
Yang H, Shang D, Xu Y, Zhang C, Feng L, Sun Z, et al. The LncRNA connectivity map: using LncRNA signatures to connect small molecules, LncRNAs, and diseases. Sci Rep. 2017;7:6655.
Yousefi H, Maheronnaghsh M, Molaei F, Mashouri L, Reza Aref A, Momeny M, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953–74.
Acknowledgements
This work was supported by Korea Brain Research Institute (KBRI) basic research program through KBRI funded by the Ministry of Science and ICT (20-BR-02-13), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1F1A1059595). All figures were created with Biorender.com.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, S., Lim, KH., Kim, SH. et al. Molecular landscape of long noncoding RNAs in brain disorders. Mol Psychiatry 26, 1060–1074 (2021). https://doi.org/10.1038/s41380-020-00947-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00947-5
This article is cited by
-
Roles of HOTAIR Long Non-coding RNA in Gliomas and Other CNS Disorders
Cellular and Molecular Neurobiology (2024)
-
The NONRATT023402.2/rno-miR-3065-5p/NGFR axis affects levodopa-induced dyskinesia in a rat model of Parkinson’s disease
Cell Death Discovery (2023)
-
Harnessing deep learning into hidden mutations of neurological disorders for therapeutic challenges
Archives of Pharmacal Research (2023)
-
Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets
Journal of Hematology & Oncology (2021)
-
TCF7L2 lncRNA: a link between bipolar disorder and body mass index through glucocorticoid signaling
Molecular Psychiatry (2021)