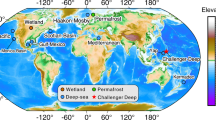
Key Points
-
The application of 'omic' approaches (for example, pyrosequencing, metagenomics, metatranscriptomics and metaproteomics) has generated unprecedented insight into Antarctic microorganisms and revealed intriguing properties about communities that can be linked to their Antarctic-specific habitats.
-
Community composition and ecosystem function are controlled by the polar light regime, biotic and abiotic environmental factors, limnological history and seed populations, biogeography and the limits of aeolian and advective dispersal caused by physical barriers and distance between sites, and perturbation caused by ecosystem change.
-
The polar austral summer is characterized by continuous high solar irradiance, which stimulates phototrophic growth and kinetically accelerates growth. Such communities tend to be oriented towards maximizing the effectiveness of light energy while switching to light-independent processes (for example, chemolithoautotrophy, phagotrophy and heterotrophic utilization of storage compounds) to survive the cold, dark winter.
-
Virus–host interactions are particularly important in the Antarctic food web, in which they not only control remineralization and influence community composition but have unanticipated roles in influencing productivity cycles. Discoveries pertaining to viruses have included systems with a high diversity of novel eukaryotic viruses, phage-resistant bacteria, and archaea capable of evading, defending against and adapting to viruses.
-
Unusual biogeochemical cycles have developed as a result of communities evolving in very specific, local environments. The indigenous communities have developed a range of traits, including a hierarchical structure, low complexity, niche adaptation, clonal dominance, mixotrophy and short-circuited nutrient cycles that enhance the use and conservation of resources.
-
Specific taxa have a major influence on overall ecosystem function, with stability of those biomes being reliant on the key, specialized and fit members maintaining function and not being affected by ecosystem perturbation, particularly anthropocentric climate change and the introduction of alien species.
Abstract
The Earth's biosphere is dominated by cold environments, and the cold biosphere is dominated by microorganisms. Microorganisms in cold Southern Ocean waters are recognized for having crucial roles in global biogeochemical cycles, including carbon sequestration, whereas microorganisms in other Antarctic aquatic biomes are not as well understood. In this Review, I consider what has been learned about Antarctic aquatic microbial ecology from 'omic' studies. I assess the factors that shape the biogeography of Antarctic microorganisms, reflect on some of the unusual biogeochemical cycles that they are associated with and discuss the important roles that viruses have in controlling ecosystem function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
20 October 2015
In the above article, there were two spelling errors. The credit line for Figure 2e should read: "Image of Deep Lake courtesy of M. Milnes, Australian Antarctic Division." The acknowledgments should read: “The author is indebted to ... M. Milnes ... for providing images of Antarctic lakes...” These have now been corrected in the online version of the article. We apologize to the readers for any misunderstanding caused.
References
Wilkins, D. et al. Key microbial drivers in Antarctic aquatic environments. FEMS Microl. Rev. 37, 303–335 (2013). Useful compendium of molecular-based studies of Antarctic aquatic microorganisms.
Chown, S. L. et al. Challenges to the future conservation of the Antarctic. Science 337, 158–159 (2012).
Chown, S. L. et al. The changing form of Antarctic biodiversity. Nature 522, 431–438 (2015).
Joughin, I., Smith, B. E. & Medley, B. Marine ice sheet collapse potentially under way for the Thwaites Glacier Basin, West Antarctica. Science 344, 735–738 (2014).
Wouters, B. et al. Glacier mass loss. Dynamic thinning of glaciers on the Southern Antarctic Peninsula. Science 348, 899–903 (2015).
Sen Gupta, A. et al. Projected changes to the Southern Hemisphere Ocean and sea ice in the IPCC AR4 climate models. J. Climate 22, 3047–3078 (2009).
Capotondi, A., Alexander, M. A., Bond, N. A., Curchitser, E. N. & Scott, J. D. Enhanced upper ocean stratification with climate change in the CMIP3 models. J. Geophys. Res. 117, C04031 (2012).
McNeil, B. I. & Matear, R. J. Southern Ocean acidification: a tipping point at 450-ppm atmospheric CO2 . Proc. Natl Acad. Sci. USA 105, 18860–18864 (2008).
Le Quéré, C. et al. Saturation of the Southern Ocean CO2 sink due to recent climate change. Science 136, 1735–1738 (2007).
Frölicher, T. L. et al. Dominance of the Southern Ocean in anthropogenic carbon and heat uptake in CMIP5 models. J. Climate 28, 862–886 (2015).
Falkowski, P. Ocean science: the power of plankton. Nature 483, S17–S20 (2012).
Cary, S. C., McDonald, I. R., Barrett, J. E. & Cowan, D. A. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8, 129–138 (2010). Insightful review describing soil and rock microbial communities in the McMurdo Dry Valleys.
Cowan, D. A. & Tow, L. A. Endangered Antarctic environments. Annu. Rev. Microbiol. 58, 649–690 (2004).
Murray, A. & Grzymski, J. Diversity and genomics of Antarctic marine microorganisms. Phil. Trans. R. Soc. B 362, 2259–2271 (2007).
Laybourn-Parry, J. No place too cold. Science 324, 1521–1522 (2009).
Feller, G. & Gerday, C. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208 (2003).
Cavicchioli, R. Cold adapted Archaea. Nat. Rev. Microbiol. 4, 331–343 (2006).
Margesin, R. & Miteva, V. Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 162, 346–361 (2011).
Siddiqui, K. S. et al. Psychrophiles. Annu. Rev. Earth Planet. Sci. 41, 87–115 (2013).
De Maayer, P., Anderson, D., Cary, C. & Cowan, D. A. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517 (2014).
Jansson, J. & Tas, N. The microbial ecology of permafrost. Nat. Rev. Microbiol. 12, 414–425 (2014).
Boetius, A., Anesio, A. M., Deming, J. W., Mikucki, J. & Rapp, J. Z. Microbial ecology of the cryosphere: sea ice and glacial habitats. Nat. Rev. Microbiol. 13, 677–690 (2015).
Williams, T. J. et al. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317 (2013).
Williams, T. J. & Cavicchioli, R. Marine metaproteomics: deciphering the microbial metabolic food web. Trends Microbiol. 22, 248–260 (2014).
Rahmstorf, S. Thermohaline circulation: the current climate. Nature 421, 699 (2003).
Matsumoto, K. Radiocarbon-based circulation age of the world oceans. J. Geophys. Res. 112, C09004 (2007).
Zika, J. D., England, M. H. & Sijp, W. P. The ocean circulation in thermohaline coordinates. J. Phys. Oceanogr. 42, 708–724 (2012).
Ballarotta, M., Falahat, S., Brodeau, L. & Döös, K. On the glacial and interglacial thermohaline circulation and the associated transports of heat and freshwater. Ocean Sci. 10, 907–921 (2014).
Ghiglione, J.-F. et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl Acad. Sci. USA 109, 17633–17638 (2012). Insightful study teasing apart the roles of isolation and ocean connectivity in influencing the distribution of microbial communities in polar ocean waters.
Sul, W. J., Oliver, T. A., Ducklow, H. W., Amaral-Zettler, L. A. & Sogin, M. L. Marine bacteria exhibit a bipolar distribution. Proc. Natl Acad. Sci. USA 110, 2342–2347 (2013).
Wilkins, D., van Sebille, E., Rintoul, S. R., Lauro, F. M. & Cavicchioli, R. Advection shapes Southern Ocean microbial assemblages independent of distance and environment effects. Nat. Commun. 4, 2457 (2013). First study to demonstrate that physical transport shapes marine microbial assemblages and should therefore be considered in the ecology and biogeography of marine microorganisms.
Fuhrman, J. A. et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl Acad. Sci. USA 105, 7774–7778 (2008).
Galand, P. E., Potvin, M., Casamayor, E. O. & Lovejoy, C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. Nature 4, 564–576 (2009).
Lauro, F. M., Chastain, R. A., Blankenship, L. E., Yayanos, A. A. & Bartlett, D. H. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Env. Microbiol. 73, 838–845 (2007).
Giebel, H.-A., Brinkhoff, T., Zwisler, W., Selje, N. & Simon, M. Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ. Microbiol. 11, 2164–2178 (2009).
Sabine, C. L. et al. The oceanic sink for anthropogenic CO2 . Science 305, 367–371 (2004).
Mikaloff Fletcher, S. E. et al. Inverse estimates of anthropogenic CO2 uptake, transport, and storage by the ocean. Global Biogeochem. Cy. 20, GB2002 (2006).
Gruber, N. et al. Oceanic sources, sinks, and transport of atmospheric CO2 . Global Biogeochem. Cy. 23, GB1005 (2009).
Thomalla, S. J. et al. Phytoplankton distribution and nitrogen dynamics in the southwest Indian subtropical gyre and Southern Ocean waters. Ocean Sci. 7, 113–127 (2011).
Wilkins, D. et al. Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics. Environ. Microbiol. 15, 1318–1333 (2013).
Brown, M. V. et al. Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 8, 595 (2012).
Selje, N., Simon, M. & Brinkhoff, T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427, 445–448 (2004).
Gianoulis, T. A. et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl Acad. Sci. USA 106, 1374–1379 (2009).
Barton, A. D., Dutkiewicz, S., Flierl, G., Bragg, J. & Follows, M. J. Patterns of diversity in marine phytoplankton. Science 327, 1509–1511 (2010).
Jiang, X. et al. Functional biogeography of ocean microbes revealed through non-negative matrix factorization. PLoS ONE 7, e43866 (2012).
Thomas, M. K., Kremer, C. T., Klausmeier, C. A. & Litchman, E. A global pattern of thermal adaptation in marine phytoplankton. Science 338, 1085–1088 (2012).
Swan, B. K. et al. Prevalent genome streamlining and latitudinal divergence of surface ocean bacterioplankton. Proc. Natl Acad. Sci. USA 110, 11463–11468 (2013).
Womack, A. M., Bohannan, B. J. & Green, J. L. Biodiversity and biogeography of the atmosphere. Phil. Trans. R. Soc. B 365, 3645–3653 (2010).
Baas Becking, L. G. M. Geobiologie Of Inleiding Tot De Milieukunde (W. P. Van Stockum & Zoon, 1934) (in Dutch).
Wynn-Williams, D. D. Aerobiology and colonization in Antarctica — the BIOTAS Programme. Grana 30, 380–393 (1991).
de Wit, R. & Bouvier, T. 'Everything is everywhere, but, the environment selects'; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8, 755–758 (2006).
Martiny, J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006).
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C. & Martiny, J. B. H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012). Excellent description of the mechanisms involved in shaping the biogeography of microbial communities.
Herbold, C. W., Lee, C. K., McDonald, I. R. & Cary, S. C. Evidence of global-scale aeolian dispersal and endemism in isolated geothermal microbial communities of Antarctica. Nat. Commun. 5, 3875 (2014).
Knowlton, C., Veerapaneni, R., D'Elia, T. & Rogers, S. O. Microbial analyses of ancient ice core sections from Greenland and Antarctica. Biology (Basel) 2, 206–232 (2013).
Morgan-Kiss, R. M., Priscu, J. C., Pocock, T., Gudynaite-Savitch, L. & Huner, N. P. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 70, 222–252 (2006).
Jungblut, A. D., Lovejoy, C. & Vincent, W. F. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 4, 191–202 (2010). Interesting study rationalizing the dispersal and selection of polar-distributed cyanobacteria, which formed the foundation for subsequent metagenomic studies.
Varin, T., Lovejoy, C., Jungblut, A. D., Vincent, W. F. & Corbeil, J. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the High Arctic. Appl. Environ. Microbiol. 78, 549–559 (2012).
Gibson, J. A. E. The meromictic lakes and stratified marine basins of the Vestfold Hills, East Antarctica. Antarct. Sci. 11, 175–192 (1999).
Ng, C. et al. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J. 4, 1002–1019 (2010). First Antarctic metaproteomics study describing an almost complete metagenome-derived genome sequence and high metaproteome coverage of an ecologically important lake microorganism.
Lauro, F. M. et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 5, 879–895 (2011). First shotgun metagenomics and metaproteomics study describing the identity and functional capacity of microbial populations throughout an entire lake, including a mathematical model describing the effects of environmental parameters on key populations.
Rankin, L. M., Gibson, J. A. E., Franzmann, P. D. & Burton, H. R. The chemical stratification and microbial communities of Ace Lake, Antarctica: a review of the characteristics of a marine-derived meromictic lake. Polarforschung 66, 35–52 (1999).
Sowell, S. M. et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3, 93–105 (2009).
Powell, L. M. et al. Ecology of a novel Synechococcus clade occurring in dense populations in saline Antarctic lakes. Mar. Ecol. Prog. Ser. 291, 65–80 (2005).
Burke, C. M. & Burton, H. R. Photosynthetic bacteria in meromictic lakes and stratified fjords of the Vestfold Hills, Antarctica. Hydrobiologia 165, 13–23 (1988).
Karr, E. A., Sattley, W. M., Jung, D. O., Madigan, M. T. & Achenbach, L. A. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 69, 4910–4914 (2003).
Sattley, W. M. & Madigan, M. T. Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 72, 5562–5568 (2006).
DeMaere, M. Z. et al. High level of inter-genera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl Acad. Sci. USA 110, 16939–16944 (2013). Study demonstrating the interplay between promiscuous gene exchange and niche adaptation in shaping Antarctic microbial communities and generating a low-complexity, hierarchically structured ecosystem.
Campbell, P. J. Primary productivity of a hypersaline Antarctic lake. Mar. Freshwater Res. 29, 717–724 (1978).
Ferris, J. M. & Burton, H. R. The annual cycle of heat content and mechanical stability of hypersaline Deep Lake, Vestfold Hills, Antarctica. Hydrobiologia 165, 115–128 (1988).
Rodriguez-Valera, F., Ruiz-Berraquero, F. & Ramos-Cormenzana, A. Isolation of extreme halophiles from seawater. Appl. Environ. Microbiol. 38, 164–165 (1979).
Munson, M. A., Nedwell, D. B. & Embley, T. M. Phylogenetic diversity of archaea in sediment samples from a coastal salt marsh. Appl. Environ. Microbiol. 63, 4729–4733 (1997).
Purdy, K. J. et al. Isolation of haloarchaea that grow at low salinities. Environ. Microbiol. 6, 591–595 (2004).
Bolhuis, H. & Stal, L. J. Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. ISME J. 5, 1701–1712 (2011).
Williams, T. J. et al. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology amongst dominant haloarchaea. ISME J. 8, 1645–1658 (2014).
Murray, A. E. et al. Microbial life at −13 °C in the brine of an ice-sealed Antarctic lake. Proc. Natl Acad. Sci. USA 109, 20626–20631 (2012). Careful consideration of the role of geochemical and biological processes occurring in an unusual ice-covered lake.
Doran, P. T., Fritsen, C. H., McKay, C. P., Priscu, J. C. & Adams, E. E. Formation and character of an ancient 19-m ice cover and underlying trapped brine in an “ice-sealed” east Antarctic lake. Proc. Natl Acad. Sci. USA 100, 26–31 (2003).
Yau, S. et al. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J. 7, 1944–1961 (2013).
Yau, S. et al. Virophage control of Antarctic algal host–virus dynamics. Proc. Natl Acad. Sci. USA 108, 6163–6168 (2011). First paper to describe an ecological role for virophages, indicating that virophages regulate host–virus interactions, influence overall carbon flux and have previously unrecognized roles in diverse aquatic ecosystems.
Mikucki, J. A. & Priscu, J. C. Bacterial diversity associated with Blood Falls, a subglacial outflow from the Taylor Glacier, Antarctica. Appl. Environ. Microbiol. 73, 4029–4039. (2007).
Mikucki, J. A. et al. A contemporary microbially maintained subglacial ferrous “ocean”. Science 324, 397–400 (2009). Colourful description of a fascinating subglacial system gleaned from the analysis of a spectacular glacier outflow.
Grzymski, J. J. et al. A metagenomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 6, 1901–1915 (2012). First study using metagenomics to identify seasonal shifts in Southern Ocean communities, highlighting the role of chemolithoautotrophic microorganisms in fixing CO 2 in winter.
Williams, T. J. et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 6, 1883–1900 (2012).
Ghiglione, J. F. & Murray, A. E. Pronounced summer to winter differences and higher wintertime richness in coastal sub-Antarctic and Antarctic marine bacterioplankton. Environ. Microbiol. 14, 617–629 (2012).
Vick, T. J. & Priscu, J. C. Bacterioplankton productivity in lakes of the Taylor Valley, Antarctica, during the polar night transition. Aquat. Microb. Ecol. 68, 77–90 (2012).
Vick-Majors, T. J., Priscu, J. C. & Amaral-Zettler, L. A. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J. 8, 778–789 (2014). First pyrosequencing study to identify shifts in lake communities during the transition to polar darkness and identifying the potentially important role of chemolithoautotrophic carbon fixation.
Bielewicz, S. et al. Protist diversity in a permanently ice-covered Antarctic lake during the polar night transition. ISME J. 5, 1559–1564 (2011).
Thurman, J. et al. Microbial dynamics and flagellate grazing during transition to winter in Lakes Hoare and Bonney, Antarctica. FEMS Microbiol. Ecol. 82, 449–458 (2012).
Laybourn-Parry, J., Marshall, W. A. & Marchant, H. J. Flagellate nutritional versatility as a key to survival in two contrasting Antarctic saline lakes. Freshwater Biol. 50, 830–838 (2005).
Kepner, R. L., Wharton, R. A. & Suttle, C. A. Viruses in Antarctic lakes. Limnol. Oceanogr. 43, 1754–1761 (1998).
López-Bueno, A. et al. High diversity of the viral community from an Antarctic lake. Science 326, 858–861 (2009). First metagenome study of an Antarctic environment, identifying a high level of new unique viruses and illustrating virus dynamics associated with physical changes occurring in the lake.
Anesio, A. M. & Bellas, C. M. Are low temperature habitats hot spots of microbial evolution driven by viruses? Trends Microbiol. 19, 52–57 (2011).
Tschitschko, B. et al. Antarctic archaea-virus interactions: metaproteome-led analysis of invasion, evasion and adaptation. ISME J. 9, 2094–2107 (2015).
Hopkins, M. et al. Diversity of environmental single-stranded DNA phages revealed by PCR amplification of the partial major capsid protein. ISME J. 8, 2093–2103 (2014).
López-Bueno, A., Rastrojo, A., Peiró, R., Arenas, M. & Alcamí, A. Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol. Ecol. 24, 4812–4825 (2015). First study describing RNA viruses in an Antarctic lake, identifying the important but uncharacterized roles that RNA viruses have in Antarctic aquatic systems.
Aguirre de Cárcer, D., López-Bueno, A., Pearce, D. A. & Alcamí, A. Biodiversity and distribution of polar freshwater DNA viruses. Sci. Adv. 1, e1400127 (2015).
Cavicchioli, R. & Erdmann, S. The discovery of Antarctic RNA viruses: a new game changer. Mol. Ecol. 24, 4809–4811 (2015).
Rodriguez-Brito, B. et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 4, 739–751 (2010).
Culley, A. I., Lang, A. S. & Suttle, C. A. Metagenomic analysis of coastal RNA virus communities. Science 312, 1795–1798 (2006).
Doming, E. & Holland, J. J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–158 (1997).
Williams, T. J. et al. Defining the response of a microorganism to growth temperature that spans its full growth temperature range (-2 °C to 28 °C) using multiplex quantitative proteomics. Environ. Microbiol. 13, 2186–2203 (2011).
Cavicchioli, R. On the concept of a psychrophile. ISME J. http://dx.doi.org/10.1038/ismej.2015.160, (2015).
Jansson, J. K. & Prosser, J. I. Microbiology: the life beneath our feet. Nature 494, 40–41 (2013).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Liu, J. & Curry, J. A. Accelerated warming of the Southern Ocean and its impacts on the hydrological cycle and sea ice. Proc. Natl Acad. Sci. USA 107, 14987–14992 (2010).
Parkinson, C. L. Global sea ice coverage from satellite data: annual cycle and 35-yr trends. J. Climate 27, 9377–9382 (2014).
Böning, C. W., Dispert, A., Visbeck, M., Rintoul, S. R. & Schwarzkopf, F. U. The response of the Antarctic Circumpolar Current to recent climate change. Nat. Geo. 1, 864–869 (2008).
Fyfe, J. C. & Saenko, O. A. Human-induced change in the Antarctic Circumpolar Current. J. Climate 18, 3068–3073 (2005).
Biastoch, A., Böning, C. W., Schwarzkopf, F. U. & Lutjeharms, J. R. E. Increase in Agulhas leakage due to poleward shift of Southern Hemisphere westerlies. Nature 462, 495–498 (2009).
Meijers, A. J. S. The Southern Ocean in the Coupled Model Intercomparison Project phase 5. Phil. Trans. R. Soc. A 372, 20130296 (2014).
Weber, T. S. & Deutsch, C. Ocean nutrient ratios governed by plankton biogeography. Nature 467, 550–554 (2010).
Murphy, E. J. et al. Developing integrated models of Southern Ocean food webs: Including ecological complexity, accounting for uncertainty and the importance of scale. Prog. Oceanogr. 102, 74–92 (2012).
Wright, A. & Siegert, M. A fourth inventory of Antarctic subglacial lakes. Antarct. Sci. 24, 659–664 (2012).
Inman, M. Antarctic drilling — the plan to unlock Lake Vostok. Science 310, 611–612 (2005).
Fox, D. Lakes under the ice: Antarctica's secret garden. Nature 512, 244–246 (2014).
Schiermeier, Q. Polar drilling problems revealed. Nature 505, 463 (2014).
Rogers, S. O. et al. Ecology of subglacial Lake Vostok (Antarctica), based on metagenomic/metatranscriptomic analyses of accretion ice. Biology (Basel) 2, 629–650 (2013).
Shtarkman, Y. M. et al. Subglacial Lake Vostok (Antarctica) accretion ice contains a diverse set of sequences from aquatic, marine and sediment-inhabiting bacteria and eukarya. PLoS ONE 8, e67221 (2013).
Siegert, M. J., Makinson, K., Blake, D., Mowlem, M. & Ross, N. An assessment of deep hot-water drilling as a means to undertake direct measurement and sampling of Antarctic subglacial lakes: experience and lessons learned from the Lake Ellsworth field season 2012/13. Ann. Glaciol. 55, 59–73 (2014).
Christner, B. C. et al. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512, 310–313 (2014). First successful deep subglacial lake exploration to identify the presence and function of lake microbial communities.
Tranter, M. Biogeochemistry: microbes eat rock under ice. Nature 512, 256–257 (2014).
Priscu, J. C. et al. A microbiologically clean strategy for access to the Whillans Ice Stream subglacial environment. Antarct. Sci. 25, 637–647 (2013).
Richter, A. et al. Subglacial Lake Vostok not expected to discharge water. Geophys. Res. Lett. 41, 6772–6778 (2014).
Bell, R. E. et al. Origin and fate of Lake Vostok water frozen to the base of the East Antarctic ice sheet. Nature 416, 307–310 (2002).
Fricker, H. A., Scambos, T., Bindschadler, R. & Padman, L. An active subglacial water system in West Antarctica mapped from space. Science 315, 1544–1548 (2007).
Fisher, A. T. et al. High geothermal heat flux measured below the West Antarctic Ice Sheet. Sci. Adv. 1, e1500093 (2015).
Wingham, D. J., Siegert, M. J., Shepherd, A. & Muir, A. S. Rapid discharge connects Antarctic subglacial lakes. Nature 440, 1033–1036 (2006).
Pattyn, F. Antarctic subglacial conditions inferred from a hybrid ice sheet/ice stream model. Earth Planet. Sci. Lett. 295, 451–461 (2010).
Wadham, J. L. et al. Potential methane reservoirs beneath Antarctica. Nature 488, 633–637 (2012). Important study highlighting the potential scale of the role that subglacial Antarctic microorganisms may have in global nutrient cycles.
Acknowledgements
The author is indebted to J. Berengut and D. Smith who crafted figures, A. Hull, M. Milnes, H. Dugan and J. Mikucki for providing images of Antarctic lakes, the Landsat Image Mosaic of Antarctica (LIMA) project for making satellite images available, D. Velázquez for discussions about Antarctic viruses, and T. Kolesnikow and T. J. Williams for insightful comments on manuscript drafts. The author's Antarctic research has been supported by the Australian Research Council, the Australian Antarctic Science Program, the Gordon and Betty Moore Foundation for DNA sequencing at the J. Craig Venter Institute and the US Department of Energy for DNA sequencing at the Joint Genome Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Glossary
- Antarctic thermohaline circulation
-
Antarctic thermohaline circulation (part of 'the global conveyer belt') arises from katabatic winds — which cause water movement that cools Southern Ocean water, which sinks because of its increased density — and from sea ice formation, which results in salt exclusion, causing the sinking of dense briny water.
- Antarctic ice sheet
-
The Antarctic continent comprises the Antarctic Peninsula Ice Sheet, the West Antarctic Ice Sheet and the East Antarctic Ice Sheet, which collectively comprise more than 500 individual glaciers.
- Stratification
-
The difference in the density of water between the surface and deeper waters (for example, upper ocean stratification from the surface to a depth of ∼200 m); stratification is increasing owing to the warming of surface waters and to an increase in freshwater content in high-latitude regions.
- Endemicity
-
The extent to which isolation and natural selection affect speciation of microorganisms in a given region (in this case, within Antarctica).
- Pelagic
-
Pelagic microorganisms inhabit the open ocean, in contrast to sea-ice microorganisms, which inhabit sea ice.
- Krill
-
Crustaceans (euphausiids) that live in the open ocean, feed on phytoplankton and to some extent zooplankton, and represent an important component in the food web bridging microorganisms (for example, primary producers) and higher trophic organisms (for example, fish, whales, seals and seabirds) that depend on them for survival.
- Pyrosequencing
-
Targeted DNA sequencing of specific gene amplicons, typically regions of small subunit rRNA (SSU rRNA) genes that can subsequently be used for generating diversity estimates and phylogenetic reconstruction of microbial community composition.
- Metagenomics
-
The study of the total DNA sequences obtained from DNA extracted from an environmental sample, with random 'shotgun' sequencing providing an inventory of genes representing the organisms present within the sample.
- Metatranscriptomics
-
The study of RNA species expressed by a microbial community present within an environmental sample determined by DNA sequencing of reverse transcribed cDNA generated from the RNA.
- Metaproteomics
-
The study of the proteins represented by a community of microorganisms present in an environmental sample, with protein identifications obtained using mass spectrometry to determine the mass of peptides derived from extracted proteins.
- Microbial communities
-
All individual microbial taxa within a defined habitat.
- Advection
-
Physical transport of components (for example, biotic and abiotic matter and heat) by ocean currents.
- Biogeography
-
Distribution of biodiversity over space and time.
- SAR11 bacterial clade
-
Members of the Bacteria comprising a distinct family within the Alphaproteobacteria that are abundant and ubiquitous in marine environments, and have important roles as oligotrophic heterotrophs.
- Mosaic genomes
-
Genomes assembled from metagenome data that are typically incomplete, in comparison to closed or draft genomes of individual cultivated laboratory-grown isolates.
- Phylotypes
-
Genetic variants of a specific lineage, often used to describe subtypes of a species: for example, an operationally defined measure of phylogenetic clustering of small subunit rRNA (SSU rRNA) gene sequences or internal transcribed spacer sequences of SSU rRNA genes.
- Aeolian dispersal
-
Movement and successful establishment of organisms (in this case, microorganisms) from one location to another mediated by the wind.
- Meromictic
-
A stratified lake that contains an upper mixed layer (mixolimnion) that does not mix with the bottom stagnant anoxic layer (monimolimnion) owing to a steep density gradient (for example, pycnocline, oxycline and chemocline) separating the two layers. By contrast, in a monomictic lake, water throughout the lake (top to bottom) mixes once per year.
- Microbial populations
-
The total contingent of one taxon (for example, species) within a microbial community.
- Green sulfur bacteria
-
(GSB). GSB (Chlorobiaceae) are phototrophic primary producers that fix CO2 at low sunlight intensities and have important roles in sulfur cycling by oxidizing reduced forms of sulfur that are often made available in the system by sulfate-reducing bacteria.
- Anoxic
-
An anoxic environment lacks oxygen, such as the bottom waters of a meromictic lake, where oxygen has been depleted and only anaerobic processes occur (for example, methanogenesis).
- Biogeochemical cycles
-
The influences of both biotic and abiotic processes on the inter-conversion of chemical substances, typically cycling chemicals through oxidized and reduced forms.
- Haloarchaea
-
Heterotrophic members of the domain Archaea that require hypersaline conditions for growth.
- Benthic
-
Benthic organisms live at the bottom of a water body (for example, a lake or an ocean); these include microorganisms growing in mats on the sediment surface and within the shallow subsurface.
- Ecotype
-
A phylotype where the genetic variation manifests in a phenotypic distinction that enables colonization of a specific ecological niche.
- Sympatric speciation
-
The process leading to the evolution of new species from a single ancestral species while inhabiting the same geographic location.
- Limnology
-
The study of inland aquatic (freshwater or saline) systems, including their physical, geological, chemical and biological characteristics.
- Virophages
-
Small viruses that are deleterious to other larger viruses, but also require the larger viruses for their own propagation (for example, by gaining entry to a host cell).
- Open discovery
-
As opposed to the testing of a specific hypothesis, open discovery involves learning something new from data acquired by observation and analysis (for example, metagenome data) that is essentially unforeseeable and is often serendipitous, akin to turning over a rock to discover what lies beneath it, opening an ancient tomb to learn what secrets it holds or viewing a new world for the first time.
Rights and permissions
About this article
Cite this article
Cavicchioli, R. Microbial ecology of Antarctic aquatic systems. Nat Rev Microbiol 13, 691–706 (2015). https://doi.org/10.1038/nrmicro3549
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3549
This article is cited by
-
Deciphering the Role of WWTPs in Cold Environments as Hotspots for the Dissemination of Antibiotic Resistance Genes
Microbial Ecology (2024)
-
Genomic analyses reveal a low-temperature adapted clade in Halorubrum, a widespread haloarchaeon across global hypersaline environments
BMC Genomics (2023)
-
Population structure of an Antarctic aquatic cyanobacterium
Microbiome (2022)
-
Microbial diversity in extreme environments
Nature Reviews Microbiology (2022)
-
Microbial diversity and ecological interactions of microorganisms in the mangrove ecosystem: Threats, vulnerability, and adaptations
Environmental Science and Pollution Research (2022)