Key Points
-
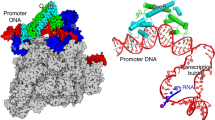
Promoter recognition in bacteria is a more complex process than originally recognized, directly involving at least five separable RNA-polymerase binding elements in the promoter and at least five promoter-binding domains or subdomains in RNA polymerase.
-
The five RNA polymerase recognition elements in bacterial promoters are the UP element that is bound by the carboxy-terminal domains of the two α subunits, the −35 element that is bound by σ region 4.2, the extended −10 element that is bound by σ region 3.0, the −10 element that is bound by σ regions 2.3 and 2.4 and the discriminator element that is bound by σ region 1.2.
-
Transcription initiation is a multistep process in which the interactions between RNA polymerase and the promoter determine the precise kinetics of the reaction. DNA-sequence variation among promoters in each of the RNA polymerase recognition elements leads to variation in the microscopic rate constants that result in the unique kinetic properties of individual promoters.
-
Regulators of transcription initiation that bind to RNA polymerase without binding to DNA include: the concentration of the first NTP in the transcript, the unusual nucleotide ppGpp, 6S RNA, anti-σ factors and the small protein DksA. In addition, the σ-factor binding protein Crl seems to increase the rate of holoenzyme assembly.
-
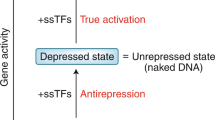
Regulators of transcription that bind to RNA polymerase without binding to DNA are able to achieve promoter-specific regulation by targeting steps in the mechanism that are rate limiting for subsets of promoters.
-
Each of these regulators can achieve global changes in the transcription profile of the cell.
Abstract
Early work identified two promoter regions, the −10 and −35 elements, that interact sequence specifically with bacterial RNA polymerase (RNAP). However, we now know that several additional promoter elements contact RNAP and influence transcription initiation. Furthermore, our picture of promoter control has evolved beyond one in which regulation results solely from activators and repressors that bind to DNA sequences near the RNAP binding site: many important transcription factors bind directly to RNAP without binding to DNA. These factors can target promoters by affecting specific kinetic steps on the pathway to open complex formation, thereby regulating RNA output from specific promoters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Record, M. T. Jr. Reznikoff, W. S., Craig, M. L., McQuade, K. L. & Schlax, P. J. in Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Neidhardt, F. C.) 792–820 (ASM, Washington DC, 1996).
Helmann, J. D. & deHaseth, P. L. Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 38, 5959–5967 (1999).
Browning, D. F. & Busby, S. J. The regulation of bacterial transcription initiation. Nature Rev. Microbiol. 2, 57–65 (2004).
Ross, W. et al. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262, 1407–1413 (1993).
Barne, K. A., Bown, J. A., Busby, S. J. & Minchin, S. D. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the 'extended-10' motif at promoters. EMBO J. 16, 4034–4040 (1997).
Haugen, S. P. et al. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell 125, 1069–1082 (2006).
Paul, B. J., Ross, W., Gaal, T. & Gourse, R. L. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38, 749–770 (2004).
Minakhin, L. et al. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl Acad. Sci. USA 98, 892–897 (2001).
Zhang, G. et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98, 811–824 (1999).
Hirata, A., Klein, B. J. & Murakami, K. S. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 (2008).
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292, 1863–1876 (2001).
Murakami, K. S., Masuda, S. & Darst, S. A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296, 1280–1284 (2002).
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002).
Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. Structural basis of transcription initiation: an RNA polymerase holoenzyme–DNA complex. Science 296, 1285–1290 (2002).
Murakami, K. S. & Darst, S. A. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13, 31–39 (2003).
Landick, R. NTP-entry routes in multi-subunit RNA polymerases. Trends Biochem. Sci. 30, 651–654 (2005).
Korzheva, N. et al. A structural model of transcription elongation. Science 289, 619–625 (2000).
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007).
Lawson, C. L. et al. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14, 10–20 (2004).
Blatter, E. E., Ross, W., Tang, H., Gourse, R. L. & Ebright, R. H. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78, 889–896 (1994).
Mekler, V. et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase–promoter open complex. Cell 108, 599–614 (2002).
Saecker, R. M. et al. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λ PR promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 319, 649–671 (2002).
Li, X. Y. & McClure, W. R. Characterization of the closed complex intermediate formed during transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 273, 23549–23557 (1998).
Cook, V. M. & deHaseth, P. L. Strand opening-deficient E. coli RNA polymerase facilitates investigation of closed complexes with promoter DNA: effects of DNA sequence and temperature. J. Biol. Chem. 282, 21319–21326 (2007).
Suh, W. C., Ross, W. & Record, M. T. Jr. Two open complexes and a requirement for Mg2+ to open the lambda PR transcription start site. Science 259, 358–361 (1993).
Naryshkin, N., Revyakin, A., Kim, Y., Mekler, V. & Ebright, R. H. Structural organization of the RNA polymerase–promoter open complex. Cell 101, 601–611 (2000).
Kontur, W. S., Saecker, R. M., Capp, M. W. & Record, M. T. Jr. Late steps in the formation of E.coli RNA polymerase — λPR promoter open complexes: characterization of conformational changes by rapid [perturbant] upshift experiments. J. Mol. Biol. 376, 1034–1047 (2008).
Davis, C. A., Bingman, C. A., Landick, R., Record, M. T. Jr & Saecker, R. M. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA 104, 7833–7838 (2007).
Sclavi, B. et al. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc. Natl Acad. Sci. USA 102, 4706–4711 (2005).
Geszvain, K. M. & Landick, R. in The Bacterial Chromosome (ed. Higgins, N. P.) 283–296 (ASM, Washington DC, 2005).
Haugen, S., Ross, W., Manrique, M. & Gourse, R. L. Fine structure of the promoter–σ region 1.2 interaction. Proc. Natl Acad. Sci. USA 105, 3292–3297 (2008).
Feklistov, A. et al. A basal promoter element recognized by free RNA polymerase σ subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell 23, 97–107 (2006).
Young, B. A., Gruber, T. M. & Gross, C. A. Views of transcription initiation. Cell 109, 417–420 (2002).
Revyakin, A., Liu, C., Ebright, R. H. & Strick, T. R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006).
Kapanidis, A. N. et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006).
Nickels, B. E. et al. The interaction between σ70 and the β-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl Acad. Sci. USA 102, 4488–4493 (2005).
Gourse, R. L., Ross, W. & Gaal, T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37, 687–695 (2000).
Ross, W., Ernst, A. & Gourse, R. L. Fine structure of E. coli RNA polymerase–promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 15, 491–506 (2001).
Lee, D. J., Busby, S. J. & Lloyd, G. S. Exploitation of a chemical nuclease to investigate the location and orientation of the Escherichia coli RNA polymerase α subunit C-terminal domains at simple promoters that are activated by cyclic AMP receptor protein. J. Biol. Chem. 278, 52944–52952 (2003).
Benoff, B. et al. Structural basis of transcription activation: the CAP-αCTD–DNA complex. Science 297, 1562–1566 (2002).
Ross, W. & Gourse, R. L. Sequence-independent upstream DNA αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl Acad. Sci. USA 102, 291–296 (2005).
Strainic, M. G. Jr, Sullivan, J. J., Velevis, A. & deHaseth, P. L. Promoter recognition by Escherichia coli RNA polymerase: effects of the UP element on open complex formation and promoter clearance. Biochemistry 37, 18074–18080 (1998).
Davis, C. A., Capp, M. W., Record, M. T. Jr & Saecker, R. M. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA 102, 285–290 (2005).
Campbell, E. A. et al. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 9, 527–539 (2002).
Jain, D., Nickels, B. E., Sun, L., Hochschild, A. & Darst, S. A. Structure of a ternary transcription activation complex. Mol. Cell 13, 45–53 (2004).
Eichenberger, P., Dethiollaz, S., Buc, H. & Geiselmann, J. Structural kinetics of transcription activation at the malT promoter of Escherichia coli by UV laser footprinting. Proc. Natl Acad. Sci. USA 94, 9022–9027 (1997).
Buckle, M., Pemberton, I. K., Jacquet, M. A. & Buc, H. The kinetics of sigma subunit directed promoter recognition by E. coli RNA polymerase. J. Mol. Biol. 285, 955–964 (1999).
Ross, W., Schneider, D. A., Paul, B. J., Mertens, A. & Gourse, R. L. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the α C-terminal domain and σ region 4. Genes Dev. 17, 1293–1307 (2003).
Chen, H., Tang, H. & Ebright, R. H. Functional interaction between RNA polymerase α subunit C-terminal domain and σ70 in UP-element- and activator-dependent transcription. Mol. Cell 11, 1621–1633 (2003).
Mitchell, J. E., Zheng, D., Busby, S. J. & Minchin, S. D. Identification and analysis of 'extended −10' promoters in Escherichia coli. Nucleic Acids Res. 31, 4689–4695 (2003).
Thouvenot, B., Charpentier, B. & Branlant, C. The strong efficiency of the Escherichia coli gapA P1 promoter depends on a complex combination of functional determinants. Biochem. J. 383, 371–382 (2004).
Hook-Barnard, I., Johnson, X. B. & Hinton, D. M. Escherichia coli RNA polymerase recognition of a σ70-dependent promoter requiring a −35 DNA element and an extended −10 TGn Motif. J. Bacteriol. 188, 8352–8359 (2006).
Camacho, A. & Salas, M. Effect of mutations in the “extended −10” motif of three Bacillus subtilis σ A–RNA polymerase-dependent promoters. J. Mol. Biol. 286, 683–693 (1999).
Voskuil, M. I. & Chambliss, G. H. The TRTGn motif stabilizes the transcription initiation open complex. J. Mol. Biol. 322, 521–532 (2002).
Hengge-Aronis, R. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5, 591–595 (2002).
Matlock, D. L. & Heyduk, T. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry 39, 12274–12283 (2000).
Young, B. A., Gruber, T. M. & Gross, C. A. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science 303, 1382–1384 (2004).
Roberts, C. W. & Roberts, J. W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell 86, 495–501 (1996).
Marr, M. T. & Roberts, J. W. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science 276, 1258–1260 (1997).
Fenton, M. S., Lee, S. J. & Gralla J. D. Escherichia coli promoter opening and −10 recognition: mutational analysis of σ70. EMBO J. 19, 1130–1137 (2000).
Qiu, J. & Helmann, J. D. Adenines at −11, −9 and −8 play a key role in the binding of Bacillus subtilis EσA RNA polymerase to −10 region single-stranded DNA. Nucleic Acids Res. 27, 4541–4546 (1999).
Tomsic, M. et al. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli σ70 in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase–promoter complexes. J. Biol. Chem. 276, 31891–31896 (2001).
Lim, H. M., Lee, H. J., Roy, S. & Adhya, S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl Acad. Sci. USA 98, 14849–14852 (2001).
Tsujikawa, L., Strainic, M. G., Watrob, H., Barkley, M. D. & DeHaseth, P. L. RNA polymerase alters the mobility of an A-residue crucial to polymerase-induced melting of promoter DNA. Biochemistry 41, 15334–15341 (2002).
Lee, H. J., Lim, H. M. & Adhya, S. An unsubstituted C2 hydrogen of adenine is critical and sufficient at the −11 position of a promoter to signal base pair deformation. J. Biol. Chem. 279, 16899–16902 (2004).
Heyduk, E., Kuznedelov, K., Severinov, K. & Heyduk, T. A consensus adenine at position −11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J. Biol. Chem. 281, 12362–12369 (2006).
McClure, W. R., Hawley, D. K., Youderian, P. & Susskind, M. M. DNA determinants of promoter selectivity in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 47 (Pt 1), 477–481 (1983).
Travers, A. A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J. Bacteriol. 141, 973–976 (1980).
Travers, A. A. & Burgess, R. R. Cyclic re-use of the RNA polymerase sigma factor. Nature 222, 537–540 (1969).
Bar-Nahum, G. & Nudler, E. Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell 106, 443–451 (2001).
Mukhopadhyay, J. et al. Translocation of σ70 with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106, 453–463 (2001).
Mooney, R. A., Darst, S. A. & Landick, R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol. Cell 20, 335–345 (2005).
Raffaelle, M., Kanin, E. I., Vogt, J., Burgess, R. R. & Ansari, A. Z. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell 20, 357–366 (2005).
Reppas, N. B., Wade, J. T., Church, G. M. & Struhl, K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell 24, 747–757 (2006).
Shimamoto, N., Kamigochi, T. & Utiyama, H. Release of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase depends mainly on time elapsed after the start of initiation, not on length of product RNA. J. Biol. Chem. 261, 11859–11865 (1986).
Sharp, M. M. et al. The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 13, 3015–3026 (1999).
Gruber, T. M. et al. Binding of the initiation factor σ70 to core RNA polymerase is a multistep process. Mol. Cell 8, 21–31 (2001).
Young, B. A. et al. A coiled-coil from the RNA polymerase β′ subunit allosterically induces selective nontemplate strand binding by σ70. Cell 105, 935–944 (2001).
Leibman, M. & Hochschild, A. A σ-core interaction of the RNA polymerase holoenzyme that enhances promoter escape. EMBO J. 26, 1579–1590 (2007).
Quinones, M., Kimsey, H. H., Ross, W., Gourse, R. L. & Waldor, M. K. LexA represses CTXφ transcription by blocking access of the α C-terminal domain of RNA polymerase to promoter DNA. J. Biol. Chem. 281, 39407–39412 (2006).
Ansari, A. Z., Bradner, J. E. & O'Halloran, T. V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374, 371–375 (1995).
Choy, H. E. et al. Repression and activation of promoter-bound RNA polymerase activity by Gal repressor. J. Mol. Biol. 272, 293–300 (1997).
Tagami, H. & Aiba, H. An inactive open complex mediated by an UP element at Escherichia coli promoters. Proc. Natl Acad. Sci. USA 96, 7202–7207 (1999).
Beck, L. L., Smith, T. G. & Hoover, T. R. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 15, 530–537 (2007).
Helmann, J. D. Anti-sigma factors. Curr. Opin. Microbiol. 2, 135–141 (1999).
Campbell, E. A. et al. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell 27, 793–805 (2007).
Gregory, B. D. et al. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA 101, 4554–4559 (2004).
Lambert, L. J., Wei, Y., Schirf, V., Demeler, B. & Werner, M. H. T4 AsiA blocks DNA recognition by remodeling σ70 region 4. EMBO J. 23, 2952–2962 (2004).
Zuber, P. Spx–RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186, 1911–1918 (2004).
Nakano, S., Erwin, K. N., Ralle, M. & Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55, 498–510 (2005).
Pratt, L. A. & Silhavy, T. J. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29, 1225–1236 (1998).
Typas, A., Barembruch, C., Possling, A. & Hengge, R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J. 26, 1569–1578 (2007).
Gaal, T., Mandel, M. J., Silhavy, T. J. & Gourse, R. L. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 188, 7966–7970 (2006).
Bougdour, A., Lelong, C. & Geiselmann, J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase σ subunit of RNA polymerase. J. Biol. Chem. 279, 19540–19550 (2004).
Robbe-Saule, V., Lopes, M. D., Kolb, A. & Norel, F. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 189, 2976–2987 (2007).
Robbe-Saule, V. et al. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 3983–3994 (2006).
Potrykus, K. & Cashel, M. (p)ppGpp: still Magical? Annu. Rev. Microbiol. 62, 35–51 (2008).
Takahashi, K., Kasai, K. & Ochi, K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl Acad. Sci. USA 101, 4320–4324 (2004).
Traxler, M. F., Chang, D. E. & Conway, T. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl Acad. Sci. USA 103, 2374–2379 (2006).
Durfee, T., Hansen, A. M., Zhi, H., Blattner, F. R. & Jin, D. J. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190, 1084–1096 (2008).
Eymann, C., Homuth, G., Scharf, C. & Hecker, M. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184, 2500–2520 (2002).
Barker, M. M., Gaal, T., Josaitis, C. A. & Gourse, R. L. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688 (2001).
Murray, H. D., Schneider, D. A. & Gourse, R. L. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12, 125–134 (2003).
Donahue, J. P. & Turnbough, C. L. Jr. Characterization of transcriptional initiation from promoters P1 and P2 of the pyrBI operon of Escherichia coli K12. J. Biol. Chem. 265, 19091–19099 (1990).
Mallik, P., Paul, B. J., Rutherford, S. T., Gourse, R. L. & Osuna, R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J. Bacteriol. 188, 5775–5782 (2006).
Paul, B. J. et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322 (2004).
Perederina, A. et al. Regulation through the secondary channel — structural framework for ppGpp–DksA synergism during transcription. Cell 118, 297–309 (2004).
Paul, B. J., Berkmen, M. B. & Gourse, R. L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl Acad. Sci. USA 102, 7823–7828 (2005).
Nakanishi, N. et al. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61, 194–205 (2006).
Costanzo, A. et al. Mechanism of regulation of the extracytoplasmic stress factor σE in Escherichia coli by DksA and the alarmone ppGpp. Mol. Microbiol. 67, 619–632 (2008).
Sharma, A. K. & Payne, S. M. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62, 469–479 (2006).
Choy, H. E. The study of guanosine 5′-diphosphate 3′-diphosphate-mediated transcription regulation in vitro using a coupled transcription–translation system. J. Biol. Chem. 275, 6783–6789 (2000).
Artsimovitch, I. et al. Structural basis for transcription regulation by alarmone ppGpp. Cell 117, 299–310 (2004).
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004).
Vrentas, C. E. et al. Still looking for the magic spot: the crystallographically-defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 377, 551–564 (2008).
Kasai, K. et al. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 188, 7111–7122 (2006).
Igarashi, K., Fujita, N. & Ishihama, A. Promoter selectivity of Escherichia coli RNA polymerase: omega factor is responsible for the ppGpp sensitivity. Nucleic Acids Res. 17, 8755–8765 (1989).
Vrentas, C. E., Gaal, T., Ross, W., Ebright, R. H. & Gourse, R. L. Response of RNA polymerase to ppGpp: requirement for the ω subunit and relief of this requirement by DksA. Genes Dev. 19, 2378–2387 (2005).
Opalka, N. et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114, 335–345 (2003).
Laptenko, O., Lee, J., Lomakin, I. & Borukhov, S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 22, 6322–6334 (2003).
Stepanova, E. et al. Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 189, 8772–8785 (2007).
Rutherford, S. T. et al. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 366, 1243–1257 (2007).
Magnusson, L. U., Gummesson, B., Joksimovic, P., Farewell, A. & Nystrom, T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189, 5193–5202 (2007).
Aberg, A., Shingler, V. & Balsalobre, C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67, 1223–1241 (2008).
Zhou, Y. N. & Jin, D. J. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl Acad. Sci. USA 95, 2908–2913 (1998).
Barker, M. M., Gaal, T. & Gourse, R. L. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305, 689–702 (2001).
Bernardo, L. M., Johansson, L. U., Solera, D., Skarfstad, E. & Shingler, V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of σ54-dependent transcription. Mol. Microbiol. 60, 749–764 (2006).
Magnusson, L. U., Farewell, A. & Nystrom, T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242 (2005).
Gourse, R. L., Ross, W. & Rutherford, S. T. General pathway for turning on promoters transcribed by RNA polymerases containing alternative σ factors. J. Bacteriol. 188, 4589–4591 (2006).
Gaal, T., Bartlett, M. S., Ross, W., Turnbough, C. L. Jr & Gourse, R. L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278, 2092–2097 (1997).
Walker, K. A., Mallik, P., Pratt, T. S. & Osuna, R. The Escherichia coli fis promoter is regulated by changes in the levels of its transcription initiation nucleotide CTP. J. Biol. Chem. 279, 50818–50828 (2004).
Lew, C. M. & Gralla, J. D. Nucleotide-dependent isomerization of Escherichia coli RNA polymerase. Biochemistry 43, 12660–12666 (2004).
Krasny, L. & Gourse, R. L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004).
Liu, C., Heath, L. S. & Turnbough, C. L. Jr. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8, 2904–2912 (1994).
Han, X. & Turnbough, C. L. Jr. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180, 705–713 (1998).
Qi, F. & Turnbough, C. L. Jr. Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 254, 552–565 (1995).
Tu, A. H. & Turnbough, C. L. Jr. Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J. Bacteriol. 179, 6665–6673 (1997).
Wilson, H. R., Archer, C. D., Liu, J. K. & Turnbough, C. L. Jr. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J. Bacteriol. 174, 514–524 (1992).
Wassarman, K. M. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 10, 164–168 (2007).
Wassarman, K. M. & Saecker, R. M. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314, 1601–1603 (2006).
Cavanagh, A. T., Klocko, A. D., Liu, X. & Wassarman, K. M. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol. Microbiol. 67, 1242–1256 (2008).
Cayley, S. & Record, M. T. Jr. Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein–DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J. Mol. Recognit. 17, 488–496 (2004).
Gralla, J. D. & Vargas, D. R. Potassium glutamate as a transcriptional inhibitor during bacterial osmoregulation. EMBO J. 25, 1515–1521 (2006).
Gourse, R. L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 16, 9789–9809 (1988).
Ohlsen, K. L. & Gralla, J. D. Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli ribosomal RNA rrnB P1 promoter. Mol. Microbiol. 6, 2243–2251 (1992).
Potrykus, K. et al. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 281, 15238–15248 (2006).
Laptenko, O. et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 25, 2131–2141 (2006).
Mukhopadhyay, J., Sineva, E., Knight, J., Levy, R. M. & Ebright, R. H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 14, 739–751 (2004).
Nickels, B. E. & Hochschild, A. Regulation of RNA polymerase through the secondary channel. Cell 118, 281–284 (2004).
Acknowledgements
We thank colleagues in our laboratory and other laboratories who provided valuable input on sections of the text and figures, including C. Turnbough, R. Landick, R. Ebright, A. Hochschild and T. Record, and apologize to those investigators whose work has not been cited owing to space limitations. Our work is supported by the National Institutes of Health (grant R37 GM37048).
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Template strand
-
The strand of DNA that enters the active site of RNA polymerase and is used as a guide for RNA synthesis. +1 is defined as the position where the template strand pairs with the nucleoside 5′-triphosphate that forms the 5′ end of the transcript. The RNA transcript is the reverse complement of the template strand and has the same sequence as the non-template strand.
- Footprinting
-
A biochemical assay for detecting protein binding sites on DNA. A protein is allowed to bind to end-labelled DNA, the DNA is subjected to limited enzymatic or chemical nuclease cleavage and DNA fragments are separated by polyacrylamide electrophoresis under conditions that allow single-nucleotide resolution.
- Crosslinking
-
A biochemical technique for identifying interactions between macromolecules. Typically, covalent bonds between macromolecules are induced by ultraviolet or chemical exposure.
- Promoter clearance
-
The step in transcription in which RNAP breaks its interactions with the promoter and begins productive RNA synthesis.
Rights and permissions
About this article
Cite this article
Haugen, S., Ross, W. & Gourse, R. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6, 507–519 (2008). https://doi.org/10.1038/nrmicro1912
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1912
This article is cited by
-
Bacterial redox response factors in the management of environmental oxidative stress
World Journal of Microbiology and Biotechnology (2023)
-
Diverse and unified mechanisms of transcription initiation in bacteria
Nature Reviews Microbiology (2021)
-
The transcriptional activator of the bfp operon in EPEC (PerA) interacts with the RNA polymerase alpha subunit
Scientific Reports (2021)
-
The stringent response regulator (p) ppGpp mediates virulence gene expression and survival in Erwinia amylovora
BMC Genomics (2020)
-
Mutational analysis of Escherichia coli GreA protein reveals new functional activity independent of antipause and lethal when overexpressed
Scientific Reports (2020)