Key Points
-
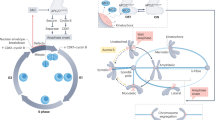
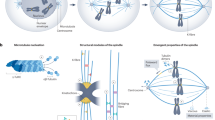
The spindle checkpoint signalling cascade prevents anaphase onset until all chromosomes are correctly attached, through their kinetochores, to spindle microtubules.
-
Molecular interactions between kinetochore and spindle checkpoint proteins have been defined and characterized.
-
There have been significant advances in understanding the molecular details of phosphoregulation and checkpoint scaffolding.
-
Monopolar spindle protein 1 (MPS1) has emerged as a direct activating kinase of the checkpoint.
-
The checkpoint response strength is variable and corresponds with the number of unattached kinetochores.
-
Inactivation of cyclin-dependent kinase 1 (CDK1) by cyclin B degradation is a basis for checkpoint inactivation during anaphase.
-
Nuclear pore complexes, in addition to kinetochores, signal the checkpoint.
Abstract
The spindle checkpoint ensures proper chromosome segregation during cell division. Unravelling checkpoint signalling has been a long-standing challenge owing to the complexity of the structures and forces that regulate chromosome segregation. New reports have now substantially advanced our understanding of checkpoint signalling mechanisms at the kinetochore, the structure that connects microtubules and chromatin. In contrast to the traditional view of a binary checkpoint response — either completely on or off — new findings indicate that the checkpoint response strength is variable. This revised perspective provides insight into how checkpoint bypass can lead to aneuploidy and informs strategies to exploit these errors for cancer treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nature Rev. Genet. 13, 189–203 (2012).
Ricke, R. M. & van Deursen, J. M. Aneuploidy in health, disease, and aging. J. Cell Biol. 201, 11–21 (2013).
McIntosh, J. R. Motors or dynamics: What really moves chromosomes? Nature Cell Biol. 14, 1234–1234 (2012).
Maresca, T. J. & Salmon, E. D. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123, 825–835 (2010).
Hoyt, M. A., Totis, L. & Roberts, B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66, 507–517 (1991).
Li, R. & Murray, A. W. Feedback control of mitosis in budding yeast. Cell 66, 519–531 (1991). References 5 and 6 established the existence of the spindle checkpoint and identified upstream checkpoint signalling genes through genetic screens.
Nicklas, R. B., Ward, S. C. & Gorbsky, G. J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 130, 929–939 (1995).
Minshull, J., S. H., Tonks, N. K. & Murray, A. W. A. MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. 79, 475–486 (1994).
Funabiki, H. & Wynne, D. J. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma 122, 135–158 (2013).
Li, X. & Nicklas, R. B. Tension-sensitive kinetochore phosphorylation and the chromosome distribution checkpoint in praying mantid spermatocytes. J. Cell Sci. 110, 537–545 (1997).
Nicklas, R. B., Campbell, M. S., Ward, S. C. & Gorbsky, G. J. Tension-sensitive kinetochore phosphorylation in vitro. J. Cell Sci. 111, 3189–3196 (1998). This classic study used innovative biophysical methods to conclusively demonstrate a role for tension in checkpoint satisfaction.
Gorbsky, G. J. & Ricketts, W. A. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J. Cell Biol. 122, 1311–1321 (1993).
Maresca, T. J. & Salmon, E. D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184, 373–381 (2009).
Foley, E. A. & Kapoor, T. M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature Rev. Mol. Cell Biol. 14, 25–37 (2012).
Vader, G., Maia, A. F. & Lens, S. M. A. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Division 3, 10 (2008).
Cheeseman, I. M., Chappie, J. S., Wilson-Kubalek, E. M. & Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 (2006). This study identified of the KMN network as a key microtubule-binding kinetochore element through biochemical reconstitution.
Varma, D. & Salmon, E. D. The KMN protein network — chief conductors of the kinetochore orchestra. J. Cell Sci. 125, 5927–5936 (2013).
Chen, R. H., Waters, J. C., Salmon, E. D. & Murray, A. W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274, 242–246 (1996). This is the first demonstration that a checkpoint protein localizes to the kinetochores, supporting the idea that the kinetochore generates the checkpoint signal.
Li, X. & Nicklas, R. B. Mitotic forces control a cell-cycle checkpoint. Nature 373, 630–632 (1995).
Rieder, C. L., Cole, R. W., Khodjakov, A. & Sluder, G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941–948 (1995). This seminal study, along with reference 19, established the kinetochore as central to the spindle checkpoint.
Weaver, B. A. A. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563 (2003).
Rieder, C. L. & Maiato, H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7, 637–651 (2004).
Dick, A. E. & Gerlich, D. W. Kinetic framework of spindle assembly checkpoint signalling. Nature Cell Biol. 15, 1370–1377 (2013). This is a clever application of established optical methods to gain unprecedented time-resolution on specific checkpoint silencing events and measure the strength of the checkpoint response.
Collin, P., Nashchekina, O., Walker, R. & Pines, J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nature Cell Biol. 15, 1378–1385 (2013). Along with references 23 and 25, this study quantitatively established the variable limits of checkpoint kinetics and duration in response to different stimuli.
Kamenz, J. & Hauf, S. Slow checkpoint activation kinetics as a safety device in anaphase. Curr. Biol. 24, 646–651 (2014).
Vázquez-Novelle, María, D. et al. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr. Biol. 24, 638–645 (2014).
Rattani, A. et al. Dependency of the spindle assembly checkpoint on Cdk1 renders the anaphase transition irreversible. Curr. Biol. 24, 630–637 (2014).
Clijsters, L. et al. Inefficient degradation of cyclin B1 re-activates the spindle checkpoint right after sister chromatid disjunction. Cell Cycle 13, 2370–2378 (2014).
Oliveira, R. A., Hamilton, R. S., Pauli, A., Davis, I. & Nasmyth, K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nature Cell Biol. 12, 185–192 (2010).
Sudakin, V. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925–936 (2001). This is the original biochemical identification of the MCC.
Burton, J. L. & Solomon, M. J. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 21, 655–667 (2007).
Chao, W. C. H., Kulkarni, K., Zhang, Z., Kong, E. H. & Barford, D. Structure of the mitotic checkpoint complex. Nature 484, 208–213 (2012). This study demonstrated the structural basis for CDC20 inactivation by the MCC.
Han, Joo, S. et al. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol. Cell 51, 92–104 (2013).
Izawa, D. & Pines, J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J. Cell Biol. 199, 27–37 (2012).
Lau, Derek, T. C. & Murray, Andrew, W. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Curr. Biol. 22, 180–190 (2012).
Luo, X. & Yu, H. Protein metamorphosis: the two-state behavior of Mad2. Structure 16, 1616–1625 (2008).
Skinner, J. J., Wood, S., Shorter, J., Englander, S. W. & Black, B. E. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. J. Cell Biol. 183, 761–768 (2008).
Howell, B. J., Hoffman, D. B., Fang, G., Murray, A. W. & Salmon, E. D. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150, 1233–1250 (2000).
Howell, B. J. et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14, 953–964 (2004). This paper describes in vivo analysis of checkpoint protein dynamics at kinetochores by FRAP. This analysis, together with those in references 38 and 40, was instrumental in revealing how the kinetochore works as a catalytic scaffold.
Shah, J. V. et al. Dynamics of centromere and kinetochore proteins: implications for checkpoint signaling and silencing. Curr. Biol. 14, 942–952 (2004).
Chen, R. H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158, 487–496 (2002).
Gillett, E. S. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164, 535–546 (2004).
Heinrich, S., Windecker, H., Hustedt, N. & Hauf, S. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J. Cell Sci. 125, 4720–4727 (2012).
Sharp-Baker, H. & Chen, R.-H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153, 1239–1250 (2001).
Vigneron, S. et al. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell 15, 4584–4596 (2004).
Kiyomitsu, T., Obuse, C. & Yanagida, M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13, 663–676 (2007). This is the initial identification of KNL1 as the BUB1 and BUBR1 kinetochore receptor. This study established that KNL1 is an essential component of the spindle checkpoint.
Kiyomitsu, T., Murakami, H. & Yanagida, M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31, 998–1011 (2011).
London, N., Ceto, S., Ranish, Jeffrey, A. & Biggins, S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22, 900–906 (2012).
Shepperd, Lindsey, A. et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22, 891–899 (2012).
Yamagishi, Y., Yang, C.-H., Tanno, Y. & Watanabe, Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nature Cell Biol. 14, 746–752 (2012). Along with references 48, 49 and 51, this study established the molecular basis for BUB1 and BUB3 localization to kinetochores, identifying a crucial kinetochore phosphorylation event in the checkpoint.
Primorac, I. et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife 2, e01030 (2013).
Campbell, L. Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. J. Cell Sci. 116, 617–628 (2003).
Suijkerbuijk, Saskia, J. E. et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell 22, 1321–1329 (2012).
Elowe, S. Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Mol. Cell. Biol. 31, 3085–3093 (2011).
Fernius, J. & Hardwick, K. G. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 3, e213 (2007).
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K.i. & Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2010).
Vleugel, M., Hoogendoorn, E., Snel, B. & Kops, Geert, J. P. L. Evolution and function of the mitotic checkpoint. Dev. Cell 23, 239–250 (2012).
Larsen, N. A., Al-Bassam, J., Wei, R. R. & Harrison, S. C. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc. Natl Acad. Sci. USA 104, 1201–1206 (2007).
Caldas, G. V. & DeLuca, J. G. KNL1: bringing order to the kinetochore. Chromosoma 123, 169–181 (2013).
Ghongane, P., Kapanidou, M., Asghar, A., Elowe, S. & Bolanos-Garcia, V. M. The dynamic protein Knl1 — a kinetochore rendezvous. J. Cell Sci. 127, 1–9 (2014).
Krenn, V., Wehenkel, A., Li, X., Santaguida, S. & Musacchio, A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196, 451–467 (2012).
Ito, D., Saito, Y. & Matsumoto, T. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proc. Natl Acad. Sci. USA 109, 209–214 (2011).
Krenn, V., Overlack, K., Primorac, I., van Gerwen, S. & Musacchio, A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT Repeats. Curr. Biol. 24, 29–39 (2014).
Vanoosthuyse, V., Valsdottir, R., Javerzat, J. P. & Hardwick, K. G. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol. Cell. Biol. 24, 9786–9801 (2004).
Johnson, V. L. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117, 1577–1589 (2004).
Kadura, S., He, X., Vanoosthuyse, V., Hardwick, K. G. & Sazer, S. The A78V mutation in the Mad3-like domain of Schizosaccharomyces pombe Bub1p perturbs nuclear accumulation and kinetochore targeting of Bub1p, Bub3p, and Mad3p and spindle assembly checkpoint function. Mol. Biol. Cell 16, 385–395 (2005).
Millband, D. N. & Hardwick, K. G. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22, 2728–2742 (2002).
D'Arcy, S., Davies, O. R., Blundell, T. L. & Bolanos-Garcia, V. M. Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J. Biol. Chem. 285, 14764–14776 (2010).
Rischitor, P. E., May, K. M. & Hardwick, K. G. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE 2, e1342 (2007).
Vleugel, M. et al. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J. Cell Biol. 203, 943–955 (2013).
Zhang, G., Lischetti, T. & Nilsson, J. A minimal number of MELT repeats supports all functions of KNL1 in chromosome segregation. J. Cell Sci. 127, 871–884 (2013).
Bolanos-Garcia, Victor, M. et al. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding site. Structure 19, 1691–1700 (2011).
Jablonski, S. A., Chan, G. K., Cooke, C. A., Earnshaw, W. C. & Yen, T. J. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107, 386–396 (1998).
Shimogawa, M. M., Wargacki, M. M., Muller, E. & Davis, T. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle 9, 3619–3628 (2010).
Kuijt, T. E. F., Omerzu, M., Saurin, A. T. & Kops, G. J. P. L. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma 123, 471–480 (2014).
Maldonado, M. & Kapoor, T. M. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nature Cell Biol. 13, 475–482 (2011). This elegant fusion study demonstrated the pivotal role of MAD1 kinetochore localization in the checkpoint.
Brady, D. M. & Hardwick, K. G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10, 675–678 (2000).
London, N. & Biggins, S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28, 140–152 (2014).
Moyle, M. W. et al. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 204, 647–657 (2014).
Kim, S., Sun, H., Tomchick, D. R., Yu, H. & Luo, X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl Acad. Sci. USA 109, 6549–6554 (2012).
Kops, G. J. P. L. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169, 49–60 (2005).
Varma, D. et al. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202, 735–746 (2013).
Kasuboski, J. M. et al. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Mol. Biol. Cell 22, 3318–3330 (2011).
Yamamoto, T. G., Watanabe, S., Essex, A. & Kitagawa, R. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183, 187–194 (2008).
Jia, L., Kim, S. & Yu, H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 38, 302–311 (2013).
Brady, D. M., Hardwick, K. G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10, 675–678 (2000).
Vink, M. et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16, 755–766 (2006).
Yang, M. et al. Insights into Mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric Mad2 dimer. PLoS Biol. 6, e50 (2008).
Chen, R.-H., Brady, D. M., Smith, D., Murray, A. W. & Hardwick, K. G. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10, 2607–2618 (1999).
Heinrich, S. et al. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 15, 291–298 (2014).
Kruse, T. et al. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep. 15, 282–290 (2014).
Ballister, E. R., Riegman, M. & Lampson, M. A. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J. Cell Biol. 204, 901–908 (2014).
Klebig, C., Korinth, D. & Meraldi, P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185, 841–858 (2009).
Kops, G. J. P. L. & Shah, J. V. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma 121, 509–525 (2012).
Jin, F. & Wang, Y. The signaling network that silences the spindle assembly checkpoint upon the establishment of chromosome bipolar attachment. Proc. Natl Acad. Sci. USA 110, 21036–21041 (2013).
Wang, Y., Jin, F., Higgins, R. & McKnight, K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle 13, 1694–1701 (2014).
Howell, B. J. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159–1172 (2001).
Barisic, M. & Geley, S. Spindly switch controls anaphase: Spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle 10, 449–456 (2011).
Gassmann, R. et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22, 2385–2399 (2008).
Gassmann, R. et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24, 957–971 (2010).
Matson, D. R., Demirel, P. B., Stukenberg, P. T. & Burke, D. J. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26, 542–547 (2012).
Matson, D. R. & Stukenberg, P. T. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 205, 541–554 (2014).
Jelluma, N., Dansen, T. B., Sliedrecht, T., Kwiatkowski, N. P. & Kops, G. J. P. L. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J. Cell Biol. 191, 281–290 (2010).
Cairo, Lucas, V., Ptak, C. & Wozniak, R. W. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol. Cell 49, 109–120 (2012).
Pinsky, B. A., Nelson, C. R. & Biggins, S. Protein Phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19, 1182–1187 (2009). Along with reference 106, this study showed that PP1 activity is essential for checkpoint silencing.
Vanoosthuyse, V. & Hardwick, K. G. A novel Protein Phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19, 1176–1181 (2009).
Rosenberg, Jessica, S., Cross, Frederick, R. & Funabiki, H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21, 942–947 (2011).
Meadows, John, C. et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and Kinesin-8 motors. Dev. Cell 20, 739–750 (2011).
Wei, R., Ngo, B., Wu, G. & Lee, W.-H. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol. Biol. Cell 22, 3584–3594 (2011).
Liu, D. et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188, 809–820 (2010).
Mirchenko, L. & Uhlmann, F. Sli15/INCENP dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 20, 1396–1401 (2010).
Vázquez-Novelle, María, D., Mirchenko, L., Uhlmann, F. & Petronczki, M. The 'anaphase problem': how to disable the mitotic checkpoint when sisters split. Biochem. Soc. Trans. 38, 1660–1666 (2010).
Palframan, W. J. Anaphase inactivation of the spindle checkpoint. Science 313, 680–684 (2006).
Hewitt, L. et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 190, 25–34 (2010).
Santaguida, S., Tighe, A., D'Alise, A. M., Taylor, S. S. & Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 (2010).
Saurin, A. T., van der Waal, M. S., Medema, R. H., Lens, S. M. A. & Kops, G. J. P. L. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nature Commun. 2, 316 (2011).
Nijenhuis, W. et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201, 217–231 (2013).
Zhu, T. et al. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J. Biol. Chem. 288, 36149–36159 (2013).
Espeut, J., Cheerambathur, D. K., Krenning, L., Oegema, K. & Desai, A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196, 469–482 (2012).
Qi, W. & Yu, H. KEN-Box-dependent degradation of the Bub1 spindle checkpoint kinase by the Anaphase-promoting Complex/Cyclosome. J. Biol. Chem. 282, 3672–3679 (2006).
Rodriguez-Bravo, V. et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 156, 1017–1031 (2014). This study identified nuclear pore-associated MAD1 as the source of the kinetochore-independent mitotic timer.
Maciejowski, J. et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190, 89–100 (2010).
Malureanu, L. A. et al. BubR1 N terminus acts as a soluble inhibitor of Cyclin B degradation by APC/CCdc20 in interphase. Dev. Cell 16, 118–131 (2009).
Meraldi, P., Draviam, V. M. & Sorger, P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45–60 (2004).
Schweizer, N. et al. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J. Cell Biol. 203, 883–893 (2013).
Cairo, L. V., Ptak, C. & Wozniak, R. W. Dual personality of Mad1: Regulation of nuclear import by a spindle assembly checkpoint protein. Nucleus 4, 367–373 (2013).
Kops, G. J. P. L., Weaver, B. A. A. & Cleveland, D. W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nature Rev. Cancer 5, 773–785 (2005).
Manchado, E., Guillamot, M. & Malumbres, M. Killing cells by targeting mitosis. Cell Death Differ. 19, 369–377 (2012).
Heinrich, S. et al. Determinants of robustness in spindle assembly checkpoint signalling. Nature Cell Biol. 15, 1328–1339 (2013).
Di Fiore, B. & Pines, J. How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 190, 501–509 (2010).
Asbury, C. L., Tien, J. F. & Davis, T. N. Kinetochores' gripping feat: conformational wave or biased diffusion? Trends Cell Biol. 21, 38–46 (2011).
Tanaka, T. U. Kinetochore–microtubule interactions: steps towards bi-orientation. EMBO J. 29, 4070–4082 (2010).
Lesage, B., Qian, J. & Bollen, M. Spindle checkpoint silencing: PP1 rips the balance. Curr. Biol. 21, R898–R903 (2011).
Musacchio, A. & Salmon, E. D. The spindle-assembly checkpoint in space and time. Nature Rev. Mol. Cell Biol. 8, 379–393 (2007).
Clute, P. & Pines, J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell Biol. 1, 82–87 (1999).
Rieder, C. L. et al. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl Acad. Sci. USA 94, 5107–5112 (1997).
Parry, D. H., Hickson, G. R. X. & O'Farrell, P. H. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr. Biol. 13, 647–653 (2003).
Jeganathan, K. B., Malureanu, L. & van Deursen, J. M. The Rae1–Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438, 1036–1039 (2005).
Schuyler, S. C., Wu, Y. F. & Kuan, V. J. W. The Mad1-Mad2 balancing act — a damaged spindle checkpoint in chromosome instability and cancer. J. Cell Sci. 125, 4197–4206 (2012).
Miniowitz-Shemtov, S. et al. Role of phosphorylation of Cdc20 in p31comet-stimulated disassembly of the mitotic checkpoint complex. Proc. Natl Acad. Sci. USA 109, 8056–8060 (2012).
Pinsky, B. A., Kung, C., Shokat, K. M. & Biggins, S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nature Cell Biol. 8, 78–83 (2006).
van der Waal, M. S. et al. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13, 847–854 (2012).
Liu, X. & Winey, M. The MPS1 family of protein kinases. Annu. Rev. Biochem. 81, 561–585 (2012).
Hardwick, K. G., Weiss, E., Luca, F. C., Winey, M. & Murray, A. W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. 273, 953–956 (1996). This study demonstrated that MPS1 kinase activity is sufficient for checkpoint activation, indicating that it is the pivotal kinase in the signalling cascade.
Maure, J.-F., Kitamura, E. & Tanaka, T. U. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 17, 2175–2182 (2007).
Tipton, A. R. et al. Monopolar Spindle 1 (MPS1) kinase promotes production of closed MAD2 (C-MAD2) conformer and assembly of the mitotic checkpoint complex. J. Biol. Chem. 288, 35149–35158 (2013).
Zich, J. et al. Kinase activity of fission yeast Mph1 is required for Mad2 and Mad3 to stably bind the Anaphase Promoting Complex. Curr. Biol. 22, 296–301 (2012).
Tipton, A. R. et al. BUBR1 and Closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J. Biol. Chem. 286, 21173–21179 (2011).
Sironi, L. et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. EMBO J. 21, 2496–2506 (2002).
Petsalaki, E. & Zachos, G. Chk2 prevents mitotic exit when the majority of kinetochores are unattached. J. Cell Biol. 205, 339–356 (2014).
Yeh, C. W., Yu, Z. C., Chen, P. H., Cheng, Y. C. & Shieh, S. Y. Phosphorylation at threonine 288 by cell cycle checkpoint kinase 2 (CHK2) controls human monopolar spindle 1 (Mps1) kinetochore localization. J. Biol. Chem. 289, 15319–15327 (2014).
Acknowledgements
The authors are grateful to the reviewers and M. Miller for thoughtful comments on the manuscript and apologize to those who were not cited owing to space limitations. N.L. was supported by a US National Institutes of Health (NIH) centre interdisciplinary training grant (T32 CA080416), and work in the Biggins laboratory is supported by NIH grants GM064386 and GM078069 to S.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Bi-oriented
-
The kinetochore–microtubule attachment state where sister kinetochores are attached exclusively to opposite spindle poles by similar numbers of microtubules.
- Cohesin complex
-
A protein complex that physically links DNA on sister or homologous chromosomes following S-phase and that must be cleaved for mitotic progression.
- E3 ubiquitin ligase
-
Enzyme responsible for transfer of ubiquitin to substrates, often targeting them for degradation by the proteasome. E3 ligases transfer ubiquitin from E2 enzymes to their substrates.
- Pseudokinase
-
A protein that is evolutionarily derived from an active kinase that has lost catalytic activity.
Rights and permissions
About this article
Cite this article
London, N., Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 15, 736–748 (2014). https://doi.org/10.1038/nrm3888
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3888
This article is cited by
-
Schizophrenia-associated Mitotic Arrest Deficient-1 (MAD1) regulates the polarity of migrating neurons in the developing neocortex
Molecular Psychiatry (2023)
-
Principles and dynamics of spindle assembly checkpoint signalling
Nature Reviews Molecular Cell Biology (2023)
-
Hyperosmotic stress-induced microtubule disassembly in Chlamydomonas reinhardtii
BMC Plant Biology (2022)
-
The cell cycle, cancer development and therapy
Molecular Biology Reports (2022)
-
The specialized mitotic behavior of human embryonic stem cells
Cell and Tissue Research (2022)