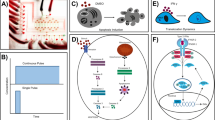
Key Points
-
This Review examines the ways in which microfluidic devices have helped to reveal the dynamics of gene regulation and intracellular signalling.
-
Gene regulatory networks often operate through highly dynamic processes that cannot be studied by stationary measurements.
-
Microfluidic devices can trap cells for long periods of time, which allows time-lapse imaging of single cells. When these devices are combined with fluorescent reporters, the time-dependent activity of a network can be measured.
-
New designs for microfludic devices allow the growth environments of cellular populations to be perturbed in non-trivial ways, such as through the creation of spatial gradients or temporal waves of chemical concentrations.
-
Mathematical models that have been created from data obtained through time-lapse fluorescence microscopy have revealed novel functions of gene networks and new regulatory pathways.
-
Multicellular and multispecies studies have also been conducted using microfluidic devices that have been designed for research in intercellular signalling.
-
It is hoped that these new technologies will eventually help to identify techniques that can more accurately model genetic regulatory networks.
Abstract
The dynamics governing gene regulation have an important role in determining the phenotype of a cell or organism. From processing extracellular signals to generating internal rhythms, gene networks are central to many time-dependent cellular processes. Recent technological advances now make it possible to track the dynamics of gene networks in single cells under various environmental conditions using microfluidic 'lab-on-a-chip' devices, and researchers are using these new techniques to analyse cellular dynamics and discover regulatory mechanisms. These technologies are expected to yield novel insights and allow the construction of mathematical models that more accurately describe the complex dynamics of gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Koide, T., Pang, W. L. & Baliga, N. S. The role of predictive modelling in rationally re-engineering biological systems. Nature Rev. Microbiol. 7, 297–305 (2009).
Reder, C. Metabolic control theory: a structural approach. J. Theor. Biol. 135, 175–201 (1988).
Edwards, J. S., Covert, M. & Palsson, B. Metabolic modelling of microbes: the flux-balance approach. Environ. Microbiol. 4, 133–140 (2002).
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protoc. 2, 2366–2382 (2007).
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002).
McDonald, J. C. et al. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 21, 27–40 (2000).
Whitesides, G. M., Ostuni, E., Takayama, S., Jiang, X. Y. & Ingber, D. E. Soft lithography in biology and biochemistry. Ann. Rev. Biomed. Eng. 3, 335–373 (2001).
Ng, J. M., Gitlin, I., Stroock, A. D. & Whitesides, G. M. Components for integrated poly(dimethylsiloxane) microfluidic systems. Electrophoresis 23, 3461–3473 (2002). References 6–8 are good reviews covering the design and manufacture of microfluidic devices.
Sia, S. K. & Whitesides, G. M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 24, 3563–3576 (2003).
Lidstrom, M. E. & Meldrum, D. R. Life-on-a-chip. Nature Rev. Microbiol. 1, 158–164 (2003).
Weibel, D. B., Diluzio, W. R. & Whitesides, G. M. Microfabrication meets microbiology. Nature Rev. Microbiol. 5, 209–218 (2007).
Chao, T. C. & Ros, A. Microfluidic single-cell analysis of intracellular compounds. J. R. Soc. Interface 5 (Suppl. 2), S139–S150 (2008).
Kim, S. M., Lee, S. H. & Suh, K. Y. Cell research with physically modified microfluidic channels: a review. Lab Chip 8, 1015–1023 (2008).
Wang, C. J. & Levchenko, A. Microfluidics technology for systems biology research. Methods Mol. Biol. 500, 203–219 (2009).
Shimomura, O., Johnson, F. H. & Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell Comp. Physiol. 59, 223–239 (1962).
Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. Creating new fluorescent probes for cell biology. Nature Rev. Mol. Cell Biol. 3, 906–918 (2002).
Shaner, N. C., Steinbach, P. A. & Tsien, R. Y. A guide to choosing fluorescent proteins. Nature Methods 2, 905–909 (2005). References 15–17 detail the properties of various fluorescent proteins that are commonly used in synthetic biology.
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Golding, I., Paulsson, J., Zawilski, S. M. & Cox, E. C. Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005).
Valencia-Burton, M., McCullough, R. M., Cantor, C. R. & Broude, N. E. RNA visualization in live bacterial cells using fluorescent protein complementation. Nature Methods 4, 421–427 (2007).
Tyagi, S. Splitting or stacking fluorescent proteins to visualize mRNA in living cells. Nature Methods 4, 391–392 (2007).
Haim, L., Zipor, G., Aronov, S. & Gerst, J. E. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nature Methods 4, 409–412 (2007).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008). This study illustrates the maturity of synthetic biology; it reports the creation of a robust and tunable synthetic gene oscillator in E. coli.
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002).
Locke, J. C. & Elowitz, M. B. Using movies to analyse gene circuit dynamics in single cells. Nature Rev. Microbiol. 7, 383–392 (2009).
Austin, D. W. et al. Gene network shaping of inherent noise spectra. Nature 439, 608–611 (2006).
Simpson, M. L., Cox, C. D. & Sayler, G. S. Frequency domain analysis of noise in autoregulated gene circuits. Proc. Natl Acad. Sci. USA 100, 4551–4556 (2003).
Cubitt, A. B. et al. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20, 448–455 (1995).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotech. 20, 87–90 (2002).
Rogers, S., Wells, R. & Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 (1986).
Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246 (1998).
Grilly, C., Stricker, J., Pang, W. L., Bennett, M. R. & Hasty, J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol. Syst. Biol. 3, 127 (2007).
Charvin, G., Cross, F. R. & Siggia, E. D. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE 3, e1468 (2008).
Khandurina, J. et al. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Anal. Chem. 72, 2995–3000 (2000).
Sanders, G. H. W. & Manz, A. Chip-based microsystems for genomic and proteomic analysis. Trends Analyt. Chem. 19, 364–378 (2000).
Lagally, E. T., Medintz, I. & Mathies, R. A. Single-molecule DNA amplification and analysis in an integrated microfluidic device. Anal. Chem. 73, 565–570 (2001).
Ramsey, J. D., Jacobson, S. C., Culbertson, C. T. & Ramsey, J. M. High-efficiency, two-dimensional separations of protein digests on microfluidic devices. Anal. Chem. 75, 3758–3764 (2003).
McClain, M. A. et al. Microfluidic devices for the high-throughput chemical analysis of cells. Anal. Chem. 75, 5646–5655 (2003).
Hong, J. W. & Quake, S. R. Integrated nanoliter systems. Nature Biotechnol. 21, 1179–1183 (2003). This review discusses the use of microfluidic devices for high-throughput biochemical assays.
Anderson, J. R. et al. Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal. Chem. 72, 3158–3164 (2000).
Chiu, D. T., Pezzoli, E., Wu, H., Stroock, A. D. & Whitesides, G. M. Using three-dimensional microfluidic networks for solving computationally hard problems. Proc. Natl Acad. Sci. USA 98, 2961–2966 (2001).
Thorsen, T., Maerkl, S. J. & Quake, S. R. Microfluidic large-scale integration. Science 298, 580–584 (2002).
Balagaddé, F. K., You, L., Hansen, C. L., Arnold, F. H. & Quake, S. R. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 309, 137–140 (2005).
Marcus, J. S., Anderson, W. F. & Quake, S. R. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 78, 3084–3089 (2006).
Maerkl, S. J. & Quake, S. R. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007).
Fu, A. Y., Spence, C., Scherer, A., Arnold, F. H. & Quake, S. R. A microfabricated fluorescence-activated cell sorter. Nature Biotech. 17, 1109–1111 (1999).
Li, P. C. H. & Harrison, D. J. Transport, manipulation, and reaction of biological cells on-chip using electrokinetic effects. Anal. Chem. 69, 1564–1568 (1997).
Fu, A. Y., Chou, H. P., Spence, C., Arnold, F. H. & Quake, S. R. An integrated microfabricated cell sorter. Anal. Chem. 74, 2451–2457 (2002).
Prokop, A. et al. NanoLiterBioReactor: long-term mammalian cell culture at nanofabricated scale. Biomed. Microdevices 6, 325–339 (2004).
Groisman, A. et al. A microfluidic chemostat for experiments with bacterial and yeast cells. Nature Methods 2, 685–689 (2005).
Cookson, S., Ostroff, N., Pang, W. L., Volfson, D. & Hasty, J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol. Syst. Biol. 1, 2005.0024 (2005).
Ryley, J. & Pereira-Smith, O. M. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast 23, 1065–1073 (2006).
Cai, L., Friedman, N. & Xie, X. S. Stochastic protein expression in individual cells at the single molecule level. Nature 440, 358–362 (2006).
Di Carlo, D., Aghdam, N. & Lee, L. P. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 78, 4925–4930 (2006).
Jeon, N. L. et al. Generation of solution and surface gradients using microfluidic systems. Langmuir 16, 8311–8316 (2000). This was one of the first investigations to use a microfluidic device capable of generating spatial chemical gradients to study a biological phenomenon.
Dertinger, S. K. W., Chiu, D. T., Jeon, N. L. & Whitesides, G. M. Generation of gradients having complex shapes using microfluidic networks. Anal. Chem. 73, 1240–1246 (2001).
Jeon, N. L. et al. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nature Biotech. 20, 826–830 (2002).
Mettetal, J. T., Muzzey, D., Gomez-Uribe, C. & van Oudenaarden, A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319, 482–484 (2008).
Hersen, P., McClean, M. N., Mahadevan, L. & Ramanathan, S. Signal processing by the HOG MAP kinase pathway. Proc. Natl Acad. Sci. USA 105, 7165–7170 (2008).
Bennett, M. R. et al. Metabolic gene regulation in a dynamically changing environment. Nature 454, 1119–1122 (2008). References 33 and 58–60 are seminal studies that used microfluidic devices to create temporal changes in the growth medium to study dynamic biological phenomena.
Siegal-Gaskins, D. & Crosson, S. Tightly regulated and heritable division control in single bacterial cells. Biophys. J. 95, 2063–2072 (2008).
McKnight, T. E. et al. Intracellular integration of synthetic nanostructures with viable cells for controlled biochemical manipulation. Nanotechnology 14, 551–556 (2003).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Gefen, O., Gabay, C., Mumcuoglu, M., Engel, G. & Balaban, N. Q. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl Acad. Sci. USA 105, 6145–6149 (2008).
Heo, J., Thomas, K. J., Seong, G. H. & Crooks, R. M. A microfluidic bioreactor based on hydrogel-entrapped E. coli: cell viability, lysis, and intracellular enzyme reactions. Anal. Chem. 75, 22–26 (2003).
Zhang, Z. et al. Microchemostat–microbial continuous culture in a polymer-based, instrumented microbioreactor. Lab Chip 6, 906–913 (2006).
Peng, X. Y. & Li, P. C. A three-dimensional flow control concept for single-cell experiments on a microchip. 1. Cell selection, cell retention, cell culture, cell balancing, and cell scanning. Anal. Chem. 76, 5273–5281 (2004).
Schmitz, C. H. J., Rowat, A. C., Koster, S. & Weitz, D. A. Dropspots: a picoliter array in a microfluidic device. Lab Chip 9, 44–49 (2009).
Park, M. C., Hur, J. Y., Kwon, K. W., Park, S. H. & Suh, K. Y. Pumpless, selective docking of yeast cells inside a microfluidic channel induced by receding meniscus. Lab Chip 6, 988–994 (2006).
Yun, K. S. & Yoon, E. Micro/nanofluidic device for single-cell-based assay. Biomed. Microdevices 7, 35–40 (2005).
Wheeler, A. R. et al. Microfluidic device for single-cell analysis. Anal. Chem. 75, 3581–3586 (2003).
Thompson, D. M. et al. Dynamic gene expression profiling using a microfabricated living cell array. Anal. Chem. 76, 4098–4103 (2004).
Lu, H. et al. Microfluidic shear devices for quantitative analysis of cell adhesion. Anal. Chem. 76, 5257–5264 (2004).
King, K. R. et al. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 7, 77–85 (2007).
Volfson, D., Cookson, S., Hasty, J. & Tsimring, L. S. Biomechanical ordering of dense cell populations. Proc. Natl Acad. Sci. USA 105, 15346–15351 (2008).
Cho, H. et al. Self-organization in high-density bacterial colonies: efficient crowd control. PLoS Biol. 5, e302 (2007).
Hao, N. et al. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol. Cell 30, 649–656 (2008).
Mao, H., Yang, T. & Cremer, P. S. A microfluidic device with a linear temperature gradient for parallel and combinatorial measurements. J. Am. Chem. Soc. 124, 4432–4435 (2002).
Holden, M. A., Kumar, S., Castellana, E. T., Beskok, A. & Cremer, P. S. Generating fixed concentration arrays in a microfluidic device. Sens. Actuators B Chem. 92, 199–207 (2003).
Zhu, X. et al. Arrays of horizontally-oriented mini-reservoirs generate steady microfluidic flows for continuous perfusion cell culture and gradient generation. Analyst 129, 1026–1031 (2004).
Walker, G. M., Ozers, M. S. & Beebe, D. J. Cell infection within a microfluidic device using virus gradients. Sens. Actuators B Chem. 98, 347–355 (2004).
Jiang, X. et al. A general method for patterning gradients of biomolecules on surfaces using microfluidic networks. Anal. Chem. 77, 2338–2347 (2005).
Irimia, D., Geba, D. A. & Toner, M. Universal microfluidic gradient generator. Anal. Chem. 78, 3472–3477 (2006).
Mao, H., Cremer, P. S. & Manson, M. D. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl Acad. Sci. USA 100, 5449–5454 (2003).
Diao, J. et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip 6, 381–388 (2006).
Lin, F. & Butcher, E. C. T cell chemotaxis in a simple microfluidic device. Lab Chip 6, 1462–1469 (2006).
Chung, B. G. et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip 5, 401–406 (2005).
Paliwal, S. et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446, 46–51 (2007).
Lin, F. et al. Generation of dynamic temporal and spatial concentration gradients using microfluidic devices. Lab Chip 4, 164–167 (2004).
Irimia, D. et al. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip 6, 191–198 (2006).
Ingolia, N. T. & Weissman, J. S. Systems biology — reverse engineering the cell. Nature 454, 1059–1062 (2008).
Tourovskaia, A., Figueroa-Masot, X. & Folch, A. Differentiation-on-a-chip: a microfluidic platform for long-term cell culture studies. Lab Chip 5, 14–19 (2005).
Olofsson, J. et al. A chemical waveform synthesizer. Proc. Natl Acad. Sci. USA 102, 8097–8102 (2005).
Lee, P. J., Gaige, T. A. & Hung, P. J. Dynamic cell culture: a microfluidic function generator for live cell microscopy. Lab Chip 9, 164–166 (2009).
Zhang, X. & Roper, M. G. Microfluidic perfusion system for automated delivery of temporal gradients to islets of Langerhans. Anal. Chem. 81, 1162–1168 (2009).
Charvin, G., Cross, F. R. & Siggia, E. D. Forced periodic expression of G1 cyclins phase-locks the budding yeast cell cycle. Proc. Natl Acad. Sci. USA 106, 6632–6637 (2009).
Chen, D. et al. The chemistrode: a droplet-based microfluidic device for stimulation and recording with high temporal, spatial, and chemical resolution. Proc. Natl Acad. Sci. USA 105, 16843–16848 (2008).
King, K. R., Wang, S., Jayaraman, A., Yarmush, M. L. & Toner, M. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip 8, 107–116 (2008).
Taylor, R. J. et al. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc. Natl Acad. Sci. USA 106, 3758–3763 (2009).
Higgins, J. M., Eddington, D. T., Bhatia, S. N. & Mahadevan, L. Sickle cell vasoocclusion and rescue in a microfluidic device. Proc. Natl Acad. Sci. USA 104, 20496–20500 (2007).
Polinkovsky, M., Gutierrez, E., Levchenko, A. & Groisman, A. Fine temporal control of the medium gas content and acidity and on-chip generation of series of oxygen concentrations for cell cultures. Lab Chip 9, 1073–1084 (2009).
Breslauer, D. N., Lee, P. J. & Lee, L. P. Microfluidics-based systems biology. Mol. Biosyst. 2, 97–112 (2006).
Kim, H. J., Boedicker, J. Q., Choi, J. W. & Ismagilov, R. F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl Acad. Sci. USA 105, 18188–18193 (2008).
Keymer, J. E., Galajda, P., Muldoon, C., Park, S. & Austin, R. H. Bacterial metapopulations in nanofabricated landscapes. Proc. Natl Acad. Sci. USA 103, 17290–17295 (2006).
Keymer, J. E., Galajda, P., Lambert, G., Liao, D. & Austin, R. H. Computation of mutual fitness by competing bacteria. Proc. Natl Acad. Sci. USA 105, 20269–20273 (2008).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Lucchetta, E. M., Lee, J. H., Fu, L. A., Patel, N. H. & Ismagilov, R. F. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434, 1134–1138 (2005).
Cai, L., Dalal, C. K. & Elowitz, M. B. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455, 485–490 (2008).
De Jong, H. Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103 (2002).
Glass, L. & Kauffman, S. A. The logical analysis of continuous, non-linear biochemical control networks. J. Theor. Biol. 39, 103–129 (1973).
Savageau, M. A. Comparison of classical and autogenous systems of regulation in inducible operons. Nature 252, 546–549 (1974).
Mather, W., Bennett, M. R., Hasty, J. & Tsimring, L. S. Delay-induced degrade-and-fire oscillations in small genetic circuits. Phys. Rev. Lett. 102, 068105 (2009).
Tran, L. M., Rizk, M. L. & Liao, J. C. Ensemble modeling of metabolic networks. Biophys. J. 95, 5606–5617 (2008).
Kepler, T. B. & Elston, T. C. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 81, 3116–3136 (2001). References 109–111 and 114 discuss some of the best modelling techniques that are common to both systems and synthetic biology, especially those that model the dynamics and stochasticity of gene regulation.
Alon, U. An Introduction to Systems Biology (Chapman and Hall/CRC, Boca Raton, 2007).
Zamir, E. & Bastiaens, P. I. Reverse engineering intracellular biochemical networks. Nature Chem. Biol. 4, 643–647 (2008).
Hasty, J., Isaacs, F., Dolnik, M., McMillen, D. & Collins, J. J. Designer gene networks: towards fundamental cellular control. Chaos 11, 207–220 (2001).
Rao, C. V. & Arkin, A. P. Stochastic chemical kinetics and the quasi-steady-state assumption: application to the Gillespie algorithm. J. Chem. Phys. 118, 4999–5010 (2003).
Gillespie, D. T. Exact stochastic simulation of coupled chemical-reactions. J. Phys. Chem. 81, 2340–2361 (1977). This paper describes the Gillespie algorithm, which is used to simulate systems of randomly interacting chemical species and is now ubiquitously used in the synthetic biology community.
Volfson, D. et al. Origins of extrinsic variability in eukaryotic gene expression. Nature 439, 861–864 (2006).
MacDonald, N. Time lag in a model of a biochemical reaction sequence with end product inhibition. J. Theor. Biol. 67, 549–556 (1977).
Mahaffy, J. M. & Pao, C. V. Models of genetic control by repression with time delays and spatial effects. J. Math. Biol. 20, 39–57 (1984).
McAdams, H. H. & Shapiro, L. Circuit simulation of genetic networks. Science 269, 650–656 (1995).
Bratsun, D., Volfson, D., Tsimring, L. S. & Hasty, J. Delay-induced stochastic oscillations in gene regulation. Proc. Natl Acad. Sci. USA 102, 14593–14598 (2005).
Bundschuh, R., Hayot, F. & Jayaprakash, C. Fluctuations and slow variables in genetic networks. Biophys. J. 84, 1606–1615 (2003).
Bennett, M. R., Volfson, D., Tsimring, L. & Hasty, J. Transient dynamics of genetic regulatory networks. Biophys. J. 92, 3501–3512 (2007).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). References 127 and 128 are two of the earliest triumphs of synthetic biology, the construction of a genetic toggle switch and a synthetic oscillator, respectively.
Acknowledgements
We would like to thank O. Mondragon and S. Cookson for initial literature searches, and B. Baumgartner for thorough readings of the drafts. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (GMO79333).
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
FlyBase
FURTHER INFORMATION
Glossary
- Intrinsic noise
-
Random, stochastic fluctuations in gene expression caused by a small number of reactants interacting in a finite volume.
- Extrinsic noise
-
Fluctuations in gene expression that are not caused by intrinsic noise.
- Time-lapse fluorescence microscopy
-
The repeated imaging of fluorescent markers using microscopy over a period of time, thus allowing a movie of the dynamics of gene expression or signalling networks to be obtained.
- Bacterial persistence
-
Similar to antibiotic resistance, bacterial persistence is the phenomenon by which a fraction of a genetically homogeneous bacterial colony will survive antibiotic treatment but retain antibiotic sensitivity following regrowth.
- Polydimethylsiloxane
-
An optically clear organic polymer that is commonly used for soft lithography.
- Stochastic
-
Probabilistic; governed by chance.
Rights and permissions
About this article
Cite this article
Bennett, M., Hasty, J. Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet 10, 628–638 (2009). https://doi.org/10.1038/nrg2625
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2625
This article is cited by
-
A synthetic population-level oscillator in non-microfluidic environments
Communications Biology (2023)
-
Competition and evolutionary selection among core regulatory motifs in gene expression control
Nature Communications (2023)
-
Majority sensing in synthetic microbial consortia
Nature Communications (2020)
-
Role of Mitochondria in Generation of Phenotypic Heterogeneity in Yeast
Journal of the Indian Institute of Science (2020)
-
Phenomenological models as effective tools to discover cellular design principles
Archives of Microbiology (2019)