Abstract
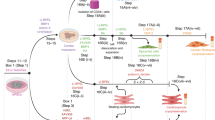
The creation of human induced pluripotent stem cells (hiPSCs) has provided an unprecedented opportunity to study tissue morphogenesis and organ development through 'organogenesis-in-a-dish'. Current approaches to cardiac organoid engineering rely on either direct cardiac differentiation from embryoid bodies (EBs) or generation of aligned cardiac tissues from predifferentiated cardiomyocytes from monolayer hiPSCs. To experimentally model early cardiac organogenesis in vitro, our protocol combines biomaterials-based cell patterning with stem cell organoid engineering. 3D cardiac microchambers are created from 2D hiPSC colonies; these microchambers approximate an early-development heart with distinct spatial organization and self-assembly. With proper training in photolithography microfabrication, maintenance of human pluripotent stem cells, and cardiac differentiation, a graduate student with guidance will likely be able to carry out this experimental protocol, which requires ∼3 weeks. We envisage that this in vitro model of human early heart development could serve as an embryotoxicity screening assay in drug discovery, regulation, and prescription for healthy fetal development. We anticipate that, when applied to hiPSC lines derived from patients with inherited diseases, this protocol can be used to study the disease mechanisms of cardiac malformations at an early stage of embryogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lancaster, M.A. & Knoblich, J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125–1247125 (2014).
Yin, X. et al. Engineering stem cell organoids. Cell Stem Cell 18, 25–38 (2016).
Willyard, C. The boom in mini stomachs, brains, breasts, kidneys and more. Nature 523, 520–522 (2015).
Simian, M. & Bissell, M.J. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 216, 31–40 (2017).
Dutta, D., Heo, I. & Clevers, H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 23, 393–410 (2017).
Ranga, A. et al. Neural tube morphogenesis in synthetic 3D microenvironments. Proc. Natl. Acad. Sci. USA 114, E3163 (2017).
Ma, Z. et al. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 6, 7413 (2015).
Mohr, J.C. et al. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials 31, 1885–1893 (2010).
Warmflash, A., Sorre, B., Etoc, F., Siggia, E.D. & Brivanlou, A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–54 (2014).
Deglincerti, A. et al. Self-organization of human embryonic stem cells on micropatterns. Nat. Protoc. 11, 2223–2232 (2016).
Fatehullah, A., Tan, S.H. & Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–54 (2016).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011).
Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013).
Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 536, 238–238 (2016).
Lancaster, M.A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Li, Y., Xu, C. & Ma, T. In vitro organogenesis from pluripotent stem cells. Organogenesis 10, 159–163 (2014).
Breckwoldt, K. et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 12, 1177–1197 (2017).
Guo, X.M. et al. Creation of engineered cardiac tissue in vitro from mouse embryonic stem cells. Circulation 113, 2229–2237 (2006).
Shkumatov, A., Baek, K. & Kong, H. Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS One 9, 1–10 (2014).
Sun, X. & Nunes, S.S. Biowire platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Methods 101, 21–6 (2016).
Wang, G. et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014).
Mathur, A. et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 5, 8883 (2015).
Ma, Z. et al. Three-dimensional filamentous human cardiac tissue model. Biomaterials 35, 1367–1377 (2014).
Hinson, J.T. et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986 (2015).
Pettinato, G., Wen, X. & Zhang, N. Formation of well-defined embryoid bodies from dissociated human induced pluripotent stem cells using microfabricated cell-repellent microwell arrays. Sci. Rep. 4, 7402 (2014).
Nunes, S.S. et al. Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat. Methods 10, 781–787 (2013).
Bergström, G., Christoffersson, J., Schwanke, K., Zweigerdt, R. & Mandenius, C.-F. Stem cell derived in vivo-like human cardiac bodies in a microfluidic device for toxicity testing by beating frequency imaging. Lab Chip 15, 3242–3249 (2015).
Matsudaira, K. et al. MEMS piezoresistive cantilever for the direct measurement of cardiomyocyte contractile force. J. Micromech. Microeng. 27, 10 (2017).
Huebsch, N. et al. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 6, 24726 (2016).
Pavesi, A. et al. Controlled electromechanical cell stimulation on-a-chip. Sci. Rep. 5, 11800 (2015).
Simmons, C.S., Petzold, B.C. & Pruitt, B.L. Microsystems for biomimetic stimulation of cardiac cells. Lab Chip 12, 3235–3248 (2012).
Caiazzo, M. et al. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 15, 344–352 (2016).
Jeon, O. & Alsberg, E. Regulation of stem cell fate in a three-dimensional micropatterned dual-crosslinked hydrogel system. Adv. Funct. Mater. 23, 4765–4775 (2013).
McBeath, R., Pirone, D.M., Nelson, C.M., Bhadriraju, K. & Chen, C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 (2004).
Musunuru, K., Domian, I.J. & Chien, K.R. Stem cell models of cardiac development and disease. Annu. Rev. Cell Dev. Biol. 26, 667–687 (2010).
Bruneau, B.G. The developmental genetics of congenital heart disease. Nature 451, 943–948 (2008).
Bressan, M. et al. Reciprocal myocardial-endocardial interactions pattern the delay in atrioventricular junction conduction. Development 141, 4149–4157 (2014).
Luxán, G. et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy 19, 193–201 (2013).
Lian, X. et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 109, E1848–E1857 (2012).
Huebsch, N. et al. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. Part C Methods 21, 467–479 (2015).
Tourovskaia, A. et al. Micropatterns of chemisorbed cell adhesion-repellent films using oxygen plasma etching and elastomeric masks. Langmuir 19, 4754–4764 (2003).
Kattman, S.J. et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240 (2011).
Burridge, P.W. et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014).
Pei, F. et al. Chemical-defined and albumin-free generation of human atrial and ventricular myocytes from human pluripotent stem cells. Stem Cell Res. 19, 94–103 (2017).
Maddah, M. et al. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Rep. 4, 621–631 (2015).
Lee, E.K., Kurokawa, Y.K., Tu, R., George, S.C. & Khine, M. Machine learning plus optical flow: a simple and sensitive method to detect cardioactive drugs. Sci. Rep. 5, 11817 (2015).
Acknowledgements
This work was supported by the Nappi Family Foundation Research Scholar Project and NIH-NIBIB R21EB021003, and in part by NIH-NCATS UH3TR000487. Z.M. acknowledges support from American Heart Association (AHA) postdoctoral fellowship 16POST27750031. P.H. acknowledges support from a National Science Foundation Integrative Graduate Education and Research Traineeship (NSF IGERT) and DMR-DGE-1068780.
Author information
Authors and Affiliations
Contributions
P.H., K.E.H., and Z.M. conceived the protocol development and finalization. P.H. and Z.M. performed biological experiments and analyzed data. J.W. contributed to the initial protocol development. B.R.C. provided the hiPSC line. P.H. wrote the manuscript with discussions and suggested improvements from all authors. Z.M. and K.E.H. funded the study, and B.R.C., K.E.H., and Z.M. supervised the project development and management.
Corresponding author
Ethics declarations
Competing interests
B.R.C. is a founder of Tenaya Therapeutics, a company focused on finding treatments for heart failure, including the use of CRISPR interference to interrogate genetic cardiomyopathies. B.R.C. holds equity in Tenaya, and Tenaya provides research support for heart failure-related research to B.R.C. K.E.H. has a financial relationship with Organos Inc., and both he and the company may benefit from commercialization of the results of this research. The other authors declare no competing financial interests.
Supplementary information
400-μm cardiac microchamber.
Video showing the beating cardiac microchamber engineered from 400-μm circle patterns, with motion vectors calculated from optical flow analysis of the motion-tracking software. (MP4 3511 kb)
600-μm cardiac microchamber.
Video showing the beating cardiac microchamber engineered from 600-μm circle patterns, with motion vectors calculated from optical flow analysis of the motion-tracking software. Reproduced with permission from ref. 7, Springer Nature. (MOV 5333 kb)
Rights and permissions
About this article
Cite this article
Hoang, P., Wang, J., Conklin, B. et al. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat Protoc 13, 723–737 (2018). https://doi.org/10.1038/nprot.2018.006
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.006
This article is cited by
-
Versatile human cardiac tissues engineered with perfusable heart extracellular microenvironment for biomedical applications
Nature Communications (2024)
-
Modeling acute myocardial infarction and cardiac fibrosis using human induced pluripotent stem cell-derived multi-cellular heart organoids
Cell Death & Disease (2024)
-
Functional precision oncology using patient-derived assays: bridging genotype and phenotype
Nature Reviews Clinical Oncology (2023)
-
Middle-out methods for spatiotemporal tissue engineering of organoids
Nature Reviews Bioengineering (2023)
-
Promises and challenges of cardiac organoids
Mammalian Genome (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.