Abstract
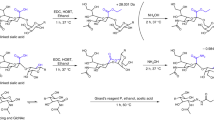
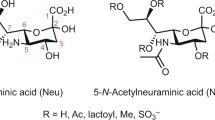
Glycosylation has a pivotal role in a diverse range of biological activities, modulating the structure and function of proteins. Glycogens coupled to the nitrogen atom (N-linked) of asparagine side chains or to the oxygen atom (O-linked) of serine and threonine side chains represent the two major protein glycosylation forms. N-glycans can be released by glycosidases, whereas O-glycans are often cleaved by chemical reaction. However, it is challenging to combine these enzymatic and chemical reactions in order to analyze both N- and O-glycans. We recently developed a glycoprotei n immobilization for glycan extraction (GIG) method that allows for the simultaneous analysis of N- and O-glycans on a solid support. GIG enables quantitative analysis of N-glycans and O-glycans from a single specimen and can be applied to a high-throughput automated platform. Here we provide a step-by-step GIG protocol that includes procedures for (i) protein immobilization on an aldehyde-active solid support by reductive amination; (ii) stabilization of fragile sialic acids by carbodiimide coupling; (iii) release of N-glycans by PNGase F digestion; (iv) release of O-glycans by β-elimination using ammonia in the presence of 1-phenyl-3-methyl-5-pyrazolone (PMP) to prevent alditol peeling from O-glycans; (v) mass spectrometry (MS) analysis; and (vi) data analysis for identification of glycans using in-house developed software (GIG Tool; free to download via http://www.biomarkercenter.org/gigtool). The GIG tool extracts precursor masses, oxonium ions and glycan fragments from tandem (liquid chromatography (LC)–MS/MS) mass spectra for glycan identification, and reporter ions from quaternary amine containing isobaric tag for glycan (QUANTITY) isobaric tags are used for quantification of the relative abundance of N-glycans. The GIG protocol takes ∼3 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Varki, A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 (1993).
Kellokumpu, S., Sormunen, R. & Kellokumpu, I. Abnormal glycosylation and altered Golgi structure in colorectal cancer: dependence on intra-Golgi pH. FEBS Lett. 516, 217–224 (2002).
Dube, D.H. & Bertozzi, C.R. Glycans in cancer and inflammation—potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 4, 477–488 (2005).
Wang, J.-Z., Grundke-Iqbal, I. & Iqbal, K. Glycosylation of microtubule-associated protein tau: An abnormal posttranslational modification in Alzheimer′s disease. Nat. Med. 2, 871–875 (1996).
Itoh, N. et al. Analysis of N-glycan in serum glycoproteins from db/db mice and humans with type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 293, E1069–E1077 (2007).
Montpetit, M.L. et al. Regulated and aberrant glycosylation modulate cardiac electrical signaling. Proc. Natl. Acad. Sci. USA 106, 16517–16522 (2009).
Yang, S. et al. Glycoproteins identified from heart failure and treatment models. Proteomics 15, 567–579 (2015).
Wang, X. et al. Overexpression of α (1, 6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 24, 935–944 (2014).
Peracaula, R., Barrabés, S., Sarrats, A., Rudd, P.M. & de Llorens, R. Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25, 207–218 (2008).
Hakomori, S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. USA 99, 10231–10233 (2002).
Hamid, U.M.A. et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology 18, 1105–1118 (2008).
Iyer, R. & Carlson, D.M. Alkaline borohydride degradation of blood group H substance. Arch. Biochem. Biophys. 142, 101–105 (1971).
Cummings, R. et al. Biosynthesis of N-and O-linked oligosaccharides of the low density lipoprotein receptor. J. Biol. Chem. 258, 15261–15273 (1983).
Takasaki, S., Mizuochi, T. & Kobata, A. Hydrazinolysis of asparagine-linked sugar chains to produce free oligosaccharides. Methods Enzymol. 83, 263–268 (1981).
Patel, T. et al. Use of hydrazine to release in intact and unreduced form both N-and O-linked oligosaccharides from glycoproteins. Biochemistry 32, 679–693 (1993).
Merry, A.H. et al. Recovery of intact 2-aminobenzamide-labeled O-glycans released from glycoproteins by hydrazinolysis. Anal. Biochem. 304, 91–99 (2002).
Zauner, G., Koeleman, C.A., Deelder, A.M. & Wuhrer, M. Mass spectrometric O-glycan analysis after combined O-glycan release by beta-elimination and 1-phenyl-3-methyl-5-pyrazolone labeling. Biochem. Biophys. Acta 1820, 1420–1428 (2012).
Huang, Y., Mechref, Y. & Novotny, M.V. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Anal. Chem. 73, 6063–6069 (2001).
Honda, S. et al. High-performance liquid chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl5-pyrazolone derivatives. Anal. Biochem. 180, 351–357 (1989).
Yang, S., Li, Y., Shah, P. & Zhang, H. Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 85, 5555–5561 (2013).
Yang, S. & Zhang, H. Glycomic analysis of glycans released from glycoproteins using chemical immobilization and mass spectrometry. Curr. Protoc. Chem. Biol. 6, 191–208 (2014).
Sekiya, S., Wada, Y. & Tanaka, K. Derivatization for stabilizing sialic acids in MALDI-MS. Anal. Chem. 77, 4962–4968 (2005).
Weiskopf, A.S., Vouros, P. & Harvey, D.J. Electrospray ionization-ion trap mass spectrometry for structural analysis of complex N-linked glycoprotein oligosaccharides. Anal. Chem. 70, 4441–4447 (1998).
Yang, S. & Zhang, H. Glycan analysis by reversible reaction to hydrazide beads and mass spectrometry. Anal. Chem. 84, 2232–2238 (2012).
Shah, P. et al. Mass spectrometric analysis of sialylated glycans with use of solid-phase labeling of sialic acids. Anal. Chem. 85, 3606–3613 (2013).
Powell, A.K. & Harvey, D.J. Stabilization of sialic acids in N-linked oligosaccharides and gangliosides for analysis by positive ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 10, 1027–1032 (1996).
Reiding, K.R., Blank, D., Kuijper, D.M., Deelder, A.M. & Wuhrer, M. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 86, 5784–5793 (2014).
Chen, P., Werner-Zwanziger, U., Wiesler, D., Pagel, M. & Novotny, M.V. Mass spectrometric analysis of benzoylated sialooligosaccharides and differentiation of terminal α2→ 3 and α2→ 6 sialogalactosylated linkages at subpicomole levels. Anal. Chem. 71, 4969–4973 (1999).
North, S.J., Hitchen, P.G., Haslam, S.M. & Dell, A. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr. Opin. Struct. Biol. 19, 498–506 (2009).
Mariño, K., Bones, J., Kattla, J.J. & Rudd, P.M. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6, 713–723 (2010).
Yang, S., Rubin, A., Eshghi, S.T. & Zhang, H. Chemoenzymatic method for glycomics: isolation, identification, and quantitation. Proteomics 16, 241–256 (2016).
Reinhold, V., Zhang, H., Hanneman, A. & Ashline, D. Toward a platform for comprehensive glycan sequencing. Mol. Cell Proteomics 12, 866–873 (2013).
Royle, L. et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376, 1–12 (2008).
Young, N.M. et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 (2002).
Lauc, G. & Wuhrer, M. High-Throughput Glycomics and Glycoproteomics: Methods and Protocols (Methods in Molecular Biology) (Humana Press, 2017).
Stavenhagen, K., Plomp, R. & Wuhrer, M. Site-specific protein N-and O-glycosylation analysis by a C18-porous graphitized carbon–liquid chromatography-electrospray ionization mass spectrometry approach using pronase treated glycopeptides. Anal. Chem. 87, 11691–11699 (2015).
Yang, S. et al. QUANTITY: an isobaric tag for quantitative glycomics. Sci. Rep. 5, 17585 (2015).
Furukawa, J et al. A versatile method for analysis of serine/threonine posttranslational modifications by β-elimination in the presence of pyrazolone analogues. Anal. Chem. 83, 9060–9067 (2011).
Huang, D., Ou, B., Hampsch-Woodill, M., Flanagan, J.A. & Prior, R.L. High-throughput assay of oxygen radical absorbance capacity (ORAC) using a multichannel liquid handling system coupled with a microplate fluorescence reader in 96-well format. J. Agric. Food Chem. 50, 4437–4444 (2002).
Chen, J., Shah, P. & Zhang, H. Solid phase extraction of N-linked glycopeptides using hydrazide tip. Anal. Chem. 85, 10670–10674 (2013).
Mach, A.J., Kim, J.H., Arshi, A., Hur, S.C. & Di Carlo, D. Automated cellular sample preparation using a centrifuge-on-a-chip. Lab Chip 11, 2827–2834 (2011).
Zhou, H., Ranish, J.A., Watts, J.D. & Aebersold, R. Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nat. Biotechnol. 20, 512–515 (2002).
Stöckmann, H., Adamczyk, B., Hayes, J. & Rudd, P.M. Automated, high-throughput IgG-antibody glycoprofiling platform. Anal. Chem. 85, 8841–8849 (2013).
Shah, P. et al. Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Mol. Cell Proteomics 14, 2753–2763 (2015).
Sun, S. et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat. Biotechnol. 34, 84–88 (2015).
Yang, S. et al. Integrated glycoprotein immobilization method for glycopeptide and glycan analysis of cardiac hypertrophy. Anal. Chem. 87, 9671–9678 (2015).
Yin, B. et al. Glycoengineering of Chinese hamster ovary cells for enhanced erythropoietin N-glycan branching and sialylation. Biotechnol. Bioeng. 112, 2343–2351 (2015).
Yang, W. et al. Glycoform analysis of recombinant and human immunodeficiency virus envelope protein gp120 via higher energy collisional dissociation and spectral-aligning strategy. Anal. Chem. 86, 6959–6967 (2014).
Chung, C.Y. et al. Integrated genome and protein editing swaps α-2, 6 sialylation for α-2, 3 sialic acid on recombinant antibodies from CHO. Biotechnol. J. 12, 1600502 (2016).
Do, D.C. et al. N-glycan in cockroach allergen regulates human basophil function. J. Allergy Clin. Immunol. 139, AB167 (2017).
Yang, S. et al. Glycan analysis by isobaric aldehyde reactive tags and mass spectrometry. Anal. Chem. 85, 8188–8195 (2013).
Lauber, M.A. et al. Rapid preparation of released N-glycans for HILIC analysis using a labeling reagent that facilitates sensitive fluorescence and ESI-MS detection. Anal. Chem. 87, 5401–5409 (2015).
Harvey, D.J. Proteomic analysis of glycosylation: structural determination of N-and O-linked glycans by mass spectrometry. Expert Rev. Proteomics 2, 87–101 (2005).
Brockhausen, I., Schachter, H. & Stanley, P. Essentials of Glycobiology 2nd edn. (Cold Spring Harbor Laboratory Press, 2009).
Wisniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359 (2009).
Morelle, W. & Michalski, J.-C. Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2, 1585–1602 (2007).
Wang, C., Fan, W., Zhang, P., Wang, Z. & Huang, L. One-pot nonreductive O-glycan release and labeling with 1-phenyl-3-methyl-5-pyrazolone followed by ESI-MS analysis. Proteomics 11, 4229–4242 (2011).
Sić, S., Maier, N.M. & Rizzi, A.M. Quantitative fingerprinting of O-linked glycans released from proteins using isotopic coded labeling with deuterated 1-phenyl-3-methyl-5-pyrazolone. J. Chromatogr. A 1408, 93–100 (2015).
Windwarder, M. & Altmann, F. Site-specific analysis of the O-glycosylation of bovine fetuin by electron-transfer dissociation mass spectrometry. J. Proteomics 108, 258–268 (2014).
Yang, S. et al. Simultaneous analyses of N-linked and O-linked glycans of ovarian cancer cells using solid-phase chemoenzymatic method. Clin. Proteomics 14, 1–12 (2017).
Acknowledgements
We thank T. Stefani and P. Shah from Johns Hopkins University for help with LC–MS and Shimadzu for providing the instrument for MALDI–MS. This work was supported by the National Institutes of Health, National Cancer Institute, the Early Detection Research Network (EDRN; U01CA152813), the Clinical Proteomic Tumor Analysis Consortium (CPTAC; U24CA160036) and the National Institutes of Health, National Heart Lung and Blood Institute Programs of Excellence in Glycosciences (P01HL107153) and the Johns Hopkins Proteomics Center (N01-HV-00240).
Author information
Authors and Affiliations
Contributions
S.Y. and H.Z. designed the research. H.Z. and L.S. directed the project. S.Y. developed the experimental protocol and conducted the experiments. S.Y. wrote the manuscript. Y.H. developed the data analysis tools and provided support with the data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Combo PDF
The Supplementary Note and Supplementary Table 1. (PDF 340 kb)
Supplementary Software 1
Format template for N-glycan database file (nlib.csv). (ZIP 164 kb)
Supplementary Software 2
Format template for O-glycan database file (olib.csv). (ZIP 424 kb)
Rights and permissions
About this article
Cite this article
Yang, S., Hu, Y., Sokoll, L. et al. Simultaneous quantification of N- and O-glycans using a solid-phase method. Nat Protoc 12, 1229–1244 (2017). https://doi.org/10.1038/nprot.2017.034
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.034
This article is cited by
-
An N-glycoproteomic site-mapping analysis reveals glycoprotein alterations in esophageal squamous cell carcinoma
Journal of Translational Medicine (2022)
-
Large-scale site-specific mapping of the O-GalNAc glycoproteome
Nature Protocols (2020)
-
Glycomics studies using sialic acid derivatization and mass spectrometry
Nature Reviews Chemistry (2020)
-
Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila
Nature Communications (2020)
-
Identification of Sialic Acid Linkages on Intact Glycopeptides via Differential Chemical Modification Using IntactGIG-HILIC
Journal of the American Society for Mass Spectrometry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.