Abstract
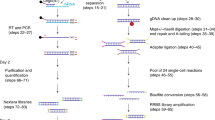
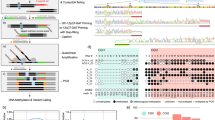
DNA methylation (DNAme) is an important epigenetic mark in diverse species. Our current understanding of DNAme is based on measurements from bulk cell samples, which obscures intercellular differences and prevents analyses of rare cell types. Thus, the ability to measure DNAme in single cells has the potential to make important contributions to the understanding of several key biological processes, such as embryonic development, disease progression and aging. We have recently reported a method for generating genome-wide DNAme maps from single cells, using single-cell bisulfite sequencing (scBS-seq), allowing the quantitative measurement of DNAme at up to 50% of CpG dinucleotides throughout the mouse genome. Here we present a detailed protocol for scBS-seq that includes our most recent developments to optimize recovery of CpGs, mapping efficiency and success rate; reduce hands-on time; and increase sample throughput with the option of using an automated liquid handler. We provide step-by-step instructions for each stage of the method, comprising cell lysis and bisulfite (BS) conversion, preamplification and adaptor tagging, library amplification, sequencing and, lastly, alignment and methylation calling. An individual with relevant molecular biology expertise can complete library preparation within 3 d. Subsequent computational steps require 1–3 d for someone with bioinformatics expertise.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Islam, S. et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21, 1160–1167 (2011).
Sasagawa, Y. et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 14, R31 (2013).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Jaitin, D.A. et al. Massively parallel single cell RNA-Seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Klein, A.M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Macosko, E.Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Clark, S.J., Lee, H.J., Smallwood, S.A., Kelsey, G. & Reik, W. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 17, 72 (2016).
Smallwood, S.A. et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014).
Farlik, M. et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 10, 1386–1397 (2015).
Gravina, S., Dong, X., Yu, B. & Vijg, J. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome Biol. 17, 150 (2016).
Nashun, B. et al. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell 60, 611–625 (2015).
Macaulay, I.C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Cokus, S.J. et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008).
Shirane, K. et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 9, e1003439 (2013).
Okae, H. et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 10, e1004868 (2014).
Fisher, S. et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011).
Li, Y. et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8, e1000533 (2010).
Bock, C. et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell 47, 633–647 (2012).
Yu, M. et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 7, 2159–2170 (2012).
Mooijman, D., Dey, S.S., Boisset, J.-C., Crosetto, N. & van Oudenaarden, A. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat. Biotechnol. 34, 852–856 (2016).
Booth, M.J. et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 8, 1841–1851 (2013).
Hu, Y. et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 17, 88 (2016).
Hou, Y. et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 26, 304–319 (2016).
Bronner, I.F., Quail, M.A., Turner, D.J. & Swerdlow, H. Improved protocols for Illumina sequencing. Curr. Protoc. Hum. Genet. 80 18.2.1 (2014).
Habibi, E. et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360–369 (2013).
Macaulay, I.C. et al. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat Protoc. 11, 2081–2103 (2016).
Guo, G. et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 6, 437–446 (2016).
von Meyenn, F. et al. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev. Cell 39, 104–115 (2016).
Eckersley-Maslin, M.A. et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 17, 179–192 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Guo, H. et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126–2135 (2013).
Acknowledgements
W.R. was supported by the Biotechnology and Biological Sciences Research Council (BBSRC), the Wellcome Trust, the EU Blueprint and EpiGeneSys. G.K. was supported by the BBSRC and the Medical Research Council (MRC). H.J.L. was supported by the EU Network of Excellence (NoE) EpiGeneSys.
Author information
Authors and Affiliations
Contributions
S.J.C. developed the updated protocol and wrote the manuscript with input from all other authors; S.A.S. and H.J.L conceived the original protocol and developed the updated version; F.K. developed the bioinformatics pipeline; and W.R. and G.K. conceived of and jointly supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
iPCRTag primer sequences. Sequence obtained is the index that is read by the Illumina machine and is the reverse complement of index portion of the primer. Primers should be ordered HPLC-purified and resuspended in EB to 100 μM. (PDF 299 kb)
Rights and permissions
About this article
Cite this article
Clark, S., Smallwood, S., Lee, H. et al. Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat Protoc 12, 534–547 (2017). https://doi.org/10.1038/nprot.2016.187
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.187
This article is cited by
-
DNA methylation remodeling and the functional implication during male gametogenesis in rice
Genome Biology (2024)
-
Characterisation and reproducibility of the HumanMethylationEPIC v2.0 BeadChip for DNA methylation profiling
BMC Genomics (2024)
-
Primordial germ cell DNA demethylation and development require DNA translesion synthesis
Nature Communications (2024)
-
Integration of multi-modal single-cell data
Nature Biotechnology (2024)
-
Dictionary learning for integrative, multimodal and scalable single-cell analysis
Nature Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.