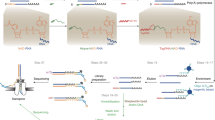
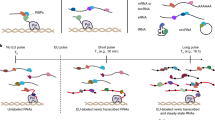
Abstract
Here we describe a protocol for NAD captureSeq that allows for the identification of nicotinamide-adenine dinucleotide (NAD)-capped RNA sequences in total RNA samples from different organisms. NAD-capped RNA is first chemo-enzymatically biotinylated with high efficiency, permitting selective capture on streptavidin beads. Then, a highly efficient library preparation protocol tailored to immobilized, 5′-modified RNA is applied, with adaptor ligation to the RNA's 3′ terminus and reverse transcription (RT) performed on-bead. Then, cDNA is released into solution, tailed, ligated to a second adaptor and PCR-amplified. After next-generation sequencing (NGS) of the DNA library, enriched sequences are identified by comparison with a control sample in which the first step of chemo-enzymatic biotinylation is omitted. Because the downstream protocol does not necessarily rely on NAD-modified but on 'clickable' or biotin-modified RNA, it can be applied to other RNA modifications or RNA–biomolecule interactions. The central part of this protocol can be completed in ∼7 d, excluding preparatory steps, sequencing and bioinformatic analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways-2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Lewis, J.D. & Izaurralde, E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247, 461–469 (1997).
Topisirovic, I., Svitkin, Y.V., Sonenberg, N. & Shatkin, A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 (2011).
Kowtoniuk, W.E., Shen, Y., Heemstra, J.M., Agarwal, I. & Liu, D.R. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl. Acad. Sci. USA 106, 7768–7773 (2009).
Chen, Y.G., Kowtoniuk, W.E., Agarwal, I., Shen, Y. & Liu, D.R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009).
Cahová, H., Winz, M.L., Höfer, K., Nübel, G. & Jäschke, A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015).
Jäschke, A., Höfer, K., Nübel, G. & Frindert, J. Cap-like structures in bacterial RNA and epitranscriptomic modification. Curr. Opin. Microbiol. 30, 44–49 (2016).
Höfer, K. et al. Structure and function of the bacterial decapping enzyme NudC. Nat. Chem. Biol. 12, 730–734 (2016).
Bird, J.G. et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016).
Höfer, K. & Jäschke, A. Molecular biology: a surprise beginning for RNA. Nature 535, 359–360 (2016).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA Biol. 7, 237–247 (2010).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 (2009).
Okada, S., Sakurai, M., Ueda, H. & Suzuki, T. Biochemical and transcriptome-wide identification of A-to-I RNA editing sites by ICE-Seq. Methods Enzymol. 560, 331–353 (2015).
Sakurai, M., Yano, T., Kawabata, H., Ueda, H. & Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 6, 733–740 (2010).
Song, C.X., Yi, C. & He, C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 30, 1107–1116 (2012).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Dominissini, D. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Tornøe, C.W., Christensen, C. & Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67, 3057–3064 (2002).
Rostovtsev, V.V., Green, L.G., Fokin, V.V. & Sharpless, K.B. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective 'ligation' of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41, 2596–2599 (2002).
Winz, M.L., Samanta, A., Benzinger, D. & Jäschke, A. Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry. Nucleic Acids Res. 40, e78 (2012).
Samanta, A., Krause, A. & Jäschke, A. A modified dinucleotide for site-specific RNA-labelling by transcription priming and click chemistry. Chem. Commun. 50, 1313–1316 (2014).
Kellner, S., Seidu-Larry, S., Burhenne, J., Motorin, Y. & Helm, M. A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA. Nucleic Acids Res. 39, 7348–7360 (2011).
Paredes, E. & Das, S.R. Click chemistry for rapid labeling and ligation of RNA. Chembiochem 12, 125–131 (2011).
Jao, C.Y. & Salic, A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784 (2008).
Clark, B.R., Halpern, R.M. & Smith, R.A. A fluorimetric method for quantitation in the picomole range of N1-methylnicotinamide and nicotinamide in serum. Anal. Biochem. 68, 54–61 (1975).
Walseth, T.F. & Lee, H.C. Synthesis and characterization of antagonists of cyclic-ADP-ribose-induced Ca2+ release. Biochim. Biophys. Acta 1178, 235–242 (1993).
Migaud, M.E., Pederick, R.L., Bailey, V.C. & Potter, B.V. Probing Aplysia californica adenosine 5′-diphosphate ribosyl cyclase for substrate binding requirements: design of potent inhibitors. Biochemistry 38, 9105–9114 (1999).
Preugschat, F., Tomberlin, G.H. & Porter, D.J. The base exchange reaction of NAD+ glycohydrolase: identification of novel heterocyclic alternative substrates. Arch. Biochem. Biophys. 479, 114–120 (2008).
Schoch, J., Staudt, M., Samanta, A., Wiessler, M. & Jäschke, A. Site-specific one-pot dual labeling of DNA by orthogonal cycloaddition chemistry. Bioconjug. Chem. 23, 1382–1386 (2012).
Winz, M.L., Linder, E.C., Andre, T., Becker, J. & Jäschke, A. Nucleotidyl transferase assisted DNA labeling with different click chemistries. Nucleic Acids Res. 43, e110 (2015).
König, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Granneman, S., Kudla, G., Petfalski, E. & Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 106, 9613–9618 (2009).
Helwak, A. & Tollervey, D. Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nat. Protoc. 9, 711–728 (2014).
Holmberg, A. et al. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 26, 501–510 (2005).
Seelig, B. & Jäschke, A. A small catalytic RNA motif with Diels-Alderase activity. Chem. Biol. 6, 167–176 (1999).
Ameta, S. & Jäschke, A. An RNA catalyst that reacts with a mechanistic inhibitor of serine proteases. Chem. Sci. 4, 957–964 (2013).
Yehudai-Resheff, S. & Schuster, G. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 28, 1139–1144 (2000).
Bard, J. et al. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science 289, 1346–1349 (2000).
Munafó, D.B. & Robb, G.B. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16, 2537–2552 (2010).
Hafner, M. et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12 (2008).
Patel, M.P., Baum, D.A. & Silverman, S.K. Improvement of DNA adenylation using T4 DNA ligase with a template strand and a strategically mismatched acceptor strand. Bioorg. Chem. 36, 46–56 (2008).
Song, Y., Liu, K.J. & Wang, T.H. Efficient synthesis of stably adenylated DNA and RNA adapters for microRNA capture using T4 RNA ligase 1. Sci. Rep. 5, 15620 (2015).
Zhelkovsky, A.M. & McReynolds, L.A. Simple and efficient synthesis of 5′ pre-adenylated DNA using thermostable RNA ligase. Nucleic Acids Res. 39, e117 (2011).
Vigneault, F., Sismour, A.M. & Church, G.M. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat. Methods 5, 777–779 (2008).
Lau, N.C., Lim, L.P., Weinstein, E.G. & Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 (2001).
Zhuang, F., Fuchs, R.T., Sun, Z., Zheng, Y. & Robb, G.B. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 40, e54 (2012).
England, T.E. & Uhlenbeck, O.C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry 17, 2069–2076 (1978).
Ohtsuka, E., Nishikawa, S., Sugiura, M. & Ikehara, M. Joining of ribooligonucleotides with T4 RNA ligase and identification of the oligonucleotide-adenylate intermediate. Nucleic Acids Res. 3, 1613–1623 (1976).
Ho, C.K., Wang, L.K., Lima, C.D. & Shuman, S. Structure and mechanism of RNA ligase. Structure 12, 327–339 (2004).
Nandakumar, J., Ho, C.K., Lima, C.D. & Shuman, S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 279, 31337–31347 (2004).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Pak, J. & Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244 (2007).
Shishkin, A.A. et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015).
Ingolia, N.T., Ghaemmaghami, S., Newman, J.R. & Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Heyer, E.E., Ozadam, H., Ricci, E.P., Cenik, C. & Moore, M.J. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 43, e2 (2015).
Schmidt, W.M. & Mueller, M.W. Controlled ribonucleotide tailing of cDNA ends (CRTC) by terminal deoxynucleotidyl transferase: a new approach in PCR-mediated analysis of mRNA sequences. Nucleic Acids Res. 24, 1789–1791 (1996).
Komura, J. & Riggs, A.D. Terminal transferase-dependent PCR: a versatile and sensitive method for in vivo footprinting and detection of DNA adducts. Nucleic Acids Res. 26, 1807–1811 (1998).
Tate, C.M. et al. Evaluation of circular DNA substrates for whole genome amplification prior to forensic analysis. Forensic Sci. Int. Genet. 6, 185–190 (2012).
Roychoudhury, R., Jay, E. & Wu, R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 3, 863–877 (1976).
Kellner, S. et al. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. 50, 3516–3518 (2014).
Van Nostrand, E.L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Zarnegar, B.J. et al. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 13, 489–492 (2016).
Martin, G. & Zavolan, M. Redesigning CLIP for efficiency, accuracy and speed. Nat. Methods 13, 482–483 (2016).
Sugimoto, Y. et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 13, R67 (2012).
Sigova, A.A. et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015).
Winz, M.L. Biological, chemical and computational investigations on RNA function and modification PhD thesis, Heidelberg University (2014).
Schulz, D., Holstein, J.M. & Rentmeister, A. A chemo-enzymatic approach for site-specific modification of the RNA cap. Angew. Chem. Int. Ed. Engl. 52, 7874–7878 (2013).
Higashida, H. et al. Sympathetic potentiation of cyclic ADP-ribose formation in rat cardiac myocytes. J. Biol. Chem. 274, 33348–33354 (1999).
Huang, F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 31, e8 (2003).
Höfer, K., Abele, F., Schlotthauer, J. & Jäschke, A. Synthesis of 5′-NAD-capped RNA. Bioconjug. Chem. 27, 874–877 (2016).
Halbritter, F., Vaidya, H.J. & Tomlinson, S.R. GeneProf: analysis of high-throughput sequencing experiments. Nat. Methods 9, 7–8 (2011).
Dumelin, C.E., Chen, Y., Leconte, A.M., Chen, Y.G. & Liu, D.R. Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 8, 913–919 (2012).
Clark, J.M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16, 9677–9686 (1988).
Zajac, P., Islam, S., Hochgerner, H., Lonnerberg, P. & Linnarsson, S. Base preferences in non-templated nucleotide incorporation by MMLV-derived reverse transcriptases. PLoS ONE 8, e85270 (2013).
Lacatena, R.M. & Cesareni, G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 294, 623–626 (1981).
Collart, M.A. & Oliviero, S. Preparation of yeast RNA. Current Protocols in Molecular Biology (eds. Ausubel F.M. et al.) Chapter 13, Unit13 12 (2001).
Nicol, J.W., Helt, G.A., Blanchard, S.G. Jr., Raja, A. & Loraine, A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731 (2009).
Speir, M.L. et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44, D717–D725 (2016).
Bustin, S.A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Acknowledgements
We thank the CellNetworks Deep Sequencing Core Facility at Heidelberg University, in particular D. Ibberson, as well as Vertis AG, in particular F. Thümmler, for Illumina sequencing and helpful discussions about adaptor and primer design. We thank J. Becker, A. Krause, A. Samanta, B. Strauß, Y.Q. Zhang, M. Tesch and other members of the Jäschke laboratory for help and discussions. M.L.W. was supported by a PhD fellowship from HBIGS. H.C. was supported by a postdoctoral fellowship from the Alexander-von-Humboldt Foundation. A.J. was supported by the German Research Council (DFG SPP 1784), the BMBF, the Helmholtz Initiative on Synthetic Biology and Baden-Württemberg Stiftung.
Author information
Authors and Affiliations
Contributions
All authors contributed to design of experiments; M.-L.W., H.C., G.N., J.F. and K.H. performed experiments; all authors analyzed experiments and contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Representative PAGE analysis of adapter ligation to the 3’-terminus of RNA.
To demonstrate the high ligation efficiency under our ligation conditions, 5 pmol of a pool of 5’-phosphorylated random 21-mer RNAs were first dephosphorylated with FastAP (Thermo Fisher Scientific) in a 10 μl reaction containing 2x standard ligation buffer, with 2-ME and BSA, but without DMSO. The dephosphorylation reaction was heat inactivated. Then, the reaction mixture was supplemented with a mixture of H2O, phosphorylated and adenylated adapter (pCTGTAGGCACCATCAAT(C3) and rAppCTGTAGGCACCATCAAT(C3)), DMSO and the respective ligases, at the same concentrations as mentioned in this article. The reactions contained either T4 RNA ligase (RNL1) alone, T4 RNA ligase 2, truncated (RNL2, tr.; New England Biolabs) alone, or both. The mixture was resolved by 12% denaturing PAGE and the gel was stained with SYBR Gold and scanned on a Typhoon scanner. Excellent ligation efficiencies were achieved with RNL1 alone or in combination with RNL2, tr., whereas non-reacted (N.R.) RNA remained when RNL2, tr. was employed alone.
Supplementary Figure 2 Product of CuAAC between ADPRC-product and Azide-PEG3-biotin.
Different parts of the resulting molecule are labelled.
Supplementary Figure 3 TdT tailing and ligation of DNA
(a) TdT tailing of single-stranded (ss) DNA1C (GATAATATGAAAGTGCAGTTTC) at varying concentrations, doped with 5’-radiolabelled DNA1C with CTP or GTP at concentrations of 12.5 mM, 1.25 mM, 125 μM, 12.5 μM and 1.25 μM for 90 min. The tailing efficiency and number of nucleotides added is far more consistent for tailing with CTP than tailing with GTP. (b) TdT tailing of ss or double-stranded (ds) DNA (DNA1C, left and DNA2 (GGAGCTCAGCCTTCACTGC), right) +/- reverse complement; 0.5 μM concentration for ss, 0.25 μM per strand for ds, each doped with 5’-radiolabelled DNA1C or DNA2). CTP concentrations were 1.25 mM (++) or 125 μM (+). Both, ssDNA and dsDNA are tailed quantitatively, although the efficiency is somewhat lower for dsDNA. (c) TdT tailing of ss DNA with varying 3’-terminal nucleotide DNA1A/C/G/T (GATAATATGAAAGTGCAGTTT(A/C/G/T)) at 5 μM concentration, doped with the respective 5’-radiolabelled DNA. Different NTPs were present at 1.25 mM concentration; 30 min reaction. The tailing efficiency does not vary substantially for different 3’-terminal nucleotides. (d), (e) Ligation of different cDNA-anchors to a random 21mer DNA (DNA3), tailed in a reaction with TdT containing either ATP, CTP, GTP or UTP, doped with the respective α-32P-NTP (Hartmann Analytic) (DNA: 0.5 μM, NTP: 125 μM, 30 min reaction). cDNA-anchors consisted of cDNA-adapter (p-CACTCGGGCACCAAGGAC-(C3)) and the respective reverse complement, with a 2 (d) or 3 (e) nt A-, C-, G-, or T-overhang. Ligation yields are indicated, as determined ratiometrically after background-subtraction using the ImageQuant software (GE Healthcare; values represent single experiments). Bands marked with an asterisk (*) results from an α-32P-GTP adduct. (f) Ligation of cDNA-anchor (as described in d, e) with 2nt G-overhang DNA (DNA1A, -C, -G, or -T) reacted with TdT and CTP, doped with α-32P-CTP (0.5 μM DNA, 125 μM CTP, 10 min reaction). Quantitative turnover is observed for all different DNAs. Thus, like TdT tailing (c) the ligation efficiency does not depend on the 3’-terminal nucleotide. All panels represent phosphor imaging scans of 15% sequencing PAGE (except (f):12% sequencing PAGE). Image panels a, b, d, f adapted with permission from ref. 68, Marie-Luise Winz; image panels c, e reproduced with permission from ref. 68, Marie-Luise Winz. Lines indicate where different parts of the same gel were combined to facilitate interpretation.
Supplementary Figure 4 Structure of ImpA with numbering scheme used to assign protons and carbon atoms in NMR.
Signals were annotated using additional C,H- and H,H-COSY data. Image reproduced with permission from ref. 68, Marie-Luise Winz.
Supplementary information
Supplementary Figures and Text
Supplementary Methods, Supplementary Notes and Supplementary Results. (PDF 807 kb)
Rights and permissions
About this article
Cite this article
Winz, ML., Cahová, H., Nübel, G. et al. Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat Protoc 12, 122–149 (2017). https://doi.org/10.1038/nprot.2016.163
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.163
This article is cited by
-
Toll/interleukin-1 receptor (TIR) domain-containing proteins have NAD-RNA decapping activity
Nature Communications (2024)
-
NADcapPro and circNC: methods for accurate profiling of NAD and non-canonical RNA caps in eukaryotes
Communications Biology (2023)
-
Identification of NAD-RNA species and ADPR-RNA decapping in Archaea
Nature Communications (2023)
-
A viral ADP-ribosyltransferase attaches RNA chains to host proteins
Nature (2023)
-
Xrn1 is a deNADding enzyme modulating mitochondrial NAD-capped RNA
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.