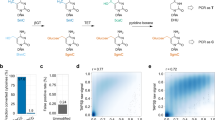
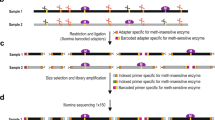
Abstract
A complete understanding of the function of the ten-eleven translocation (TET) family of dioxygenase-mediated DNA demethylation requires new methods to quantitatively map oxidized 5-methylcytosine (5mC) bases at high resolution. We have recently developed a methylase-assisted bisulfite sequencing (MAB-seq) method that allows base-resolution mapping of 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), two oxidized 5mC bases indicative of active DNA demethylation events. In standard bisulfite sequencing (BS-seq), unmodified C, 5fC and 5caC are read as thymine; thus 5fC and 5caC cannot be distinguished from C. In MAB-seq, unmodified C is enzymatically converted to 5mC, allowing direct mapping of rare modifications such as 5fC and 5caC. By combining MAB-seq with chemical reduction of 5fC to 5hmC, we also developed caMAB-seq, a method for direct 5caC mapping. Compared with subtraction-based mapping methods, MAB-seq and caMAB-seq require less sequencing effort and enable robust statistical calling of 5fC and/or 5caC. MAB-seq and caMAB-seq can be adapted to map 5fC/5caC at the whole-genome scale (WG-MAB-seq), within specific genomic regions enriched for enhancer-marking histone modifications (chromatin immunoprecipitation (ChIP)-MAB-seq), or at CpG-rich sequences (reduced-representation (RR)-MAB-seq) such as gene promoters. The full protocol, including DNA preparation, enzymatic treatment, library preparation and sequencing, can be completed within 6–8 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Smith, Z.D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Wu, H. & Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014).
Pastor, W.A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Kohli, R.M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Maiti, A. & Drohat, A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011).
Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 (2011).
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011).
Pastor, W.A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Xu, Y. et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 (2011).
Wu, H. & Zhang, Y. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell Cycle 10 2428–2436 ( 2011).
Dawlaty, M.M. et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323 (2013).
Kang, J. et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. USA 112, E4236–4245 (2015).
Gu, T.P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477 610 (2011).
Dawlaty, M.M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Koh, K.P. et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213 (2011).
Yamaguchi, S. et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447 (2012).
Yamaguchi, S., Shen, L., Liu, Y., Sendler, D. & Zhang, Y. Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464 (2013).
Zhang, R.R. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 (2013).
Rudenko, A. et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122 (2013).
Kaas, G.A. et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093 (2013).
Cimmino, L., Abdel-Wahab, O., Levine, R.L. & Aifantis, I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9, 193–204 (2011).
Ko, M. et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 263, 6–21 (2015).
Song, C.X. & He, C. Potential functional roles of DNA demethylation intermediates. Trends Biochem. Sci. 38, 480–484 (2013).
Wu, H. & Zhang, Y. Charting oxidized methylcytosines at base resolution. Nat. Struct. Mol. Biol. 22, 656–661 (2015).
Spruijt, C.G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Iurlaro, M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14, R119 (2013).
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012).
Kellinger, M.W. et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012).
Wang, L. et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 523, 621–625 (2015).
Raiber, E.A. et al. 5-Formylcytosine alters the structure of the DNA double helix. Nat. Struct. Mol. Biol. 22, 44–49 (2015).
Szulik, M.W. et al. Differential stabilities and sequence-dependent base pair opening dynamics of Watson-Crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine. Biochemistry 54, 1294–1305 (2015).
Wu, H., Wu, X., Shen, L. & Zhang, Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 32, 1231–1240 (2014).
Song, C.-X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Shen, L. et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 (2013).
Neri, F. et al. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 10, 674–683 (2015).
Guo, F. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–458 (2014).
Lister, R. et al. Global epigenomic reconfiguration during Mammalian brain development. Science 341, 1237905 (2013).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Hu, L. et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013).
Hashimoto, H. et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 (2014).
Sun, Z. et al. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell 57, 750–761 (2015).
Xia, B. et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Methods 12, 1047–1050 (2015).
Booth, M.J., Marsico, G., Bachman, M., Beraldi, D. & Balasubramanian, S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6, 435–440 (2014).
Lu, X. et al. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 25, 386–389 (2015).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Lu, F., Liu, Y., Jiang, L., Yamaguchi, S. & Zhang, Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 28, 2103–2119 (2014).
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Thorvaldsdottir, H., Robinson, J.T. & Mesirov, J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 (2013).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Liu, Y., Siegmund, K.D., Laird, P.W. & Berman, B.P. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 13, R61 (2012).
Sotiriou, S. et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat. Med. 8, 514–517 (2002).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Yin, R. et al. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. J. Am. Chem. Soc. 135, 10396–10403 (2013).
Keane, T.M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011).
Wang, K. et al. Q-RRBS: a quantitative reduced representation bisulfite sequencing method for single-cell methylome analyses. Epigenetics 10, 775–783 (2015).
Booth, M.J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Acknowledgements
H.W. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and is currently supported by the National Human Genome Research Institute (R00HG007982). X.W. was supported by the China Scholarship Council. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
H.W. and X.W. performed experiments and carried out data analysis. H.W., X.W. and Y.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Wu, H., Wu, X. & Zhang, Y. Base-resolution profiling of active DNA demethylation using MAB-seq and caMAB-seq. Nat Protoc 11, 1081–1100 (2016). https://doi.org/10.1038/nprot.2016.069
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.069
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.