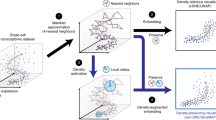
Abstract
High-throughput single-cell technologies provide an unprecedented view into cellular heterogeneity, yet they pose new challenges in data analysis and interpretation. In this protocol, we describe the use of Spanning-tree Progression Analysis of Density-normalized Events (SPADE), a density-based algorithm for visualizing single-cell data and enabling cellular hierarchy inference among subpopulations of similar cells. It was initially developed for flow and mass cytometry single-cell data. We describe SPADE's implementation and application using an open-source R package that runs on Mac OS X, Linux and Windows systems. A typical SPADE analysis on a 2.27-GHz processor laptop takes ∼5 min. We demonstrate the applicability of SPADE to single-cell RNA-seq data. We compare SPADE with recently developed single-cell visualization approaches based on the t-distribution stochastic neighborhood embedding (t-SNE) algorithm. We contrast the implementation and outputs of these methods for normal and malignant hematopoietic cells analyzed by mass cytometry and provide recommendations for appropriate use. Finally, we provide an integrative strategy that combines the strengths of t-SNE and SPADE to infer cellular hierarchy from high-dimensional single-cell data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Qiu, P. Inferring phenotypic properties from single-cell characteristics. PLoS One 7, e37038 (2012).
Levina, E. & Bickel, P. The earthmover's distance is the Mallows distance: some insights from statistics. Proceedings of ICCV 2001 (Vancouver, Canada) 251–256 (2001).
Ngom, A. et al. Pattern Recognition in Bioinformatics. Lecture Notes in Computer Science Vol. 7986 (Berlin: Springer, 2013).
Amir el, A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Aghaeepour, N. et al. RchyOptimyx: cellular hierarchy optimization for flow cytometry. Cytometry A 81, 1022–1030 (2012).
Shekhar, K. et al. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE). Proc. Natl. Acad. Sci. USA 111, 202–207 (2014).
Anchang, B. et al. CCAST: a model-based gating strategy to isolate homogeneous subpopulations in a heterogeneous population of single cells. PLoS Comput. Biol. 10, e1003664 (2014).
Zare, H. et al. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics 11, 403 (2010).
Lo, K. et al. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 10, 145 (2009).
Mosmann, T.R. et al. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, part 2: biological evaluation. Cytometry A 85, 422–433 (2014).
Pyne, S. et al. Automated high-dimensional flow cytometric data analysis. Proc. Natl. Acad. Sci. USA 106, 8519–8524 (2009).
Linderman, M., Qiu, P., Simonds, E. & Bjornson, Z. SPADE – An analysis and visualization tool for Flow Cytometry. R package version 1.20. http://bioconductor.org (2016).
Linderman, M.D. et al. CytoSPADE: high-performance analysis and visualization of high-dimensional cytometry data. Bioinformatics 28, 2400–2401 (2012).
Kotecha, N., Krutzik, P.O. & Irish, J.M. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom Chapter 10, 10.17 (2010).
Pettie, S. & Ramach, V. An optimal minimum spanning tree algorithm. JACM 49, 49–60 (1999).
Prim, C.M. Shortest connection networks and some generalizations. Bell Syst. Tech. J. 36, 1389–1401 (1957).
Zunder, E.R., Lujan, E., Goltsev, Y., Wernig, M. & Nolan, G. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell cytometry. Cell Stem Cell 16, 323–337 (2015).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
van der Maaten, L. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Qiu, P., Gentles, A.J. & Plevritis, S.K. Discovering biological progression underlying microarray samples. PLoS Comput. Biol. 7, e1001123 (2011).
Aghaeepour, N. et al. Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods 10, 228–238 (2013).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A 87A, 636–645 (2015).
Yu, M. et al. Hierarchical clustering in minimum spanning trees. Chaos 25, 023107 (2015).
Fruchterman, T.M.J. & Reingold, E.M. Graph drawing by force-directed placement Software: Practice & Experience. 21, 1129–1164 (1991).
Qiu, P. & Plevritis, S.K. TreeVis: a MATLAB-based tool for tree visualization. Comput. Methods Programs Biomed. 109, 74–76 (2013).
Kamada, T. & Kawai, S. An algorithm for drawing general undirected graphs. Inform. Process. Lett. 31, 7–15 (1989).
Acknowledgements
This study was primarily supported by National Institutes of Health (NIH) grant U54CA149145, with S.K.P. as principal investigator. G.P.N. is supported by NIH grants U19 AI057229, 1U19AI100627, U54 CA149145, N01-HV-00242, 1R01CA130826, 5R01AI073724, R01 GM109836, R01CA184968, 1R01NS089533, P01 CA034233, R33 CA183654, R33 CA183692, 41000411217, 201303028, HHSN272201200028C, HHSN272200700038C, and 5U54CA143907; CIRM DR1-01477; Department of Defense grants OC110674 and 11491122; FDA grant HHSF223201210194C; Bill and Melinda Gates Foundation grant OPP1113682; Alliance for Lupus Research grant 218518; and the Rachford and Carlota A. Harris Endowed Professorship. P.Q. is supported by NIH grant R01 CA163481. S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017-09) and NIH grant R00 GM104148-03.
Author information
Authors and Affiliations
Contributions
B.A., T.D.P.H., S.C.B., P.Q., Z.B., M.L., G.P.N. and S.K.P. contributed to the concept of SPADE analyses. B.A., T.D.P.H. and S.K.P. were involved in the concept and design of the integrated SPADE–t-SNE analysis. B.A. and T.D.P.H. performed computational analyses. All authors interpreted the results. B.A. and S.K.P. wrote the initial drafts of the manuscript. All authors edited, read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent (S10-010) for the SPADE algorithm has been applied for on behalf of Stanford University.
Integrated supplementary information
Supplementary Figure 1 SPADE analysis of normal human bone marrow trees colored by the median intensities of CD45RA for 5 different number of clusters denoted by k.
Reducing k below 100 produces a sparse tree. The trees with most cells concentrated in the branches correspond to k=100 and k=200. In general the results are not highly sensitive to the number of clusters chosen.
Supplementary Figure 2 SPADE analysis of normal human bone marrow using 5 different bootstrapping samples of the same data.
The trees are colored by all 24 subpopulations from Bendall et al. (2011)2 denoted here as subpopulations 1-24. Visual analysis of the SPADE tree branches between different runs and cluster colors will show that the branches and relative positioning of the clusters within a branch are often preserved.
Supplementary information
Combo PDF
Supplementary Figures 1 and 2 (PDF 581 kb)
Supplementary Data 1
Unlabeled subsample bone marrow data set from Bendall et al.2 used to explain the SPADE workflow in Figure 1. (ZIP 2000 kb)
Supplementary Data 2
MCM FCS file containing expression data from manually gated normal human bone marrow cells from Bendall et al.2 used for comparison analysis. MCM FCS file of ALL single-cell data from Amir et al.7 used for comparison analysis. Data in FCS file format containing the mouse lung epithelial RNA-seq expression from Treutlein et al.23. (ZIP 22244 kb)
Supplementary Data 3
MCM FCS file of ALL single-cell data from Amir et al. (2013)7 used for comparison analysis. (ZIP 18185 kb)
Supplementary Data 4
Data in FCS file format containing the Mouse lung epithelial RNA-Seq expression from Treutlein et al. (2014)23 (ZIP 4 kb)
Supplementary Software
R code for how to combine SPADE and t-SNE to generate a ‘SPADE forest’ for a single FCS file. (PDF 1511 kb)
Rights and permissions
About this article
Cite this article
Anchang, B., Hart, T., Bendall, S. et al. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc 11, 1264–1279 (2016). https://doi.org/10.1038/nprot.2016.066
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.066
This article is cited by
-
Identifying dysregulated immune cell subsets following volumetric muscle loss with pseudo-time trajectories
Communications Biology (2023)
-
Microfluidics applications for high-throughput single cell sequencing
Journal of Nanobiotechnology (2021)
-
Local delivery of FTY720 induces neutrophil activation through chemokine signaling in an oronasal fistula model
Regenerative Engineering and Translational Medicine (2021)
-
Slow progressors to type 1 diabetes lose islet autoantibodies over time, have few islet antigen-specific CD8+ T cells and exhibit a distinct CD95hi B cell phenotype
Diabetologia (2020)
-
Progenitor identification and SARS-CoV-2 infection in human distal lung organoids
Nature (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.