Abstract
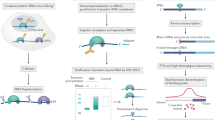
Mammalian cell nuclei contain three RNA polymerases (RNAP I, RNAP II and RNAP III), which transcribe different gene subsets, and whose active forms are contained in supramolecular complexes known as 'transcription factories.' These complexes are difficult to isolate because they are embedded in the 3D structure of the nucleus. Factories exchange components with the soluble nucleoplasmic pool over time as gene expression programs change during development or disease. Analysis of their content can provide information on the nascent transcriptome and its regulators. Here we describe a protocol for the isolation of large factory fragments under isotonic salt concentrations in <72 h. It relies on DNase I–mediated detachment of chromatin from the nuclear substructure of freshly isolated, unfixed cells, followed by caspase treatment to release multi-megadalton factory complexes. These complexes retain transcriptional activity, and isolation of their contents is compatible with downstream analyses by mass spectrometry (MS) or RNA-sequencing (RNA-seq) to catalog the proteins and RNA associated with sites of active transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Roeder, R.G. The eukaryotic transcriptional machinery: complexities and mechanisms unforeseen. Nat. Med. 9, 1239–1244 (2003).
Cramer, P. et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37, 337–352 (2008).
Das, R. et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 26, 867–881 (2007).
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 (2009).
Papantonis, A. & Cook, P.R. Transcription factories: genome organization and gene regulation. Chem. Rev. 113, 8683–8705 (2013).
Chakalova, L. & Fraser, P. Organization of transcription. Cold Spring Harb. Perspect. Biol. 2, a000729 (2010).
Sutherland, H. & Bickmore, W.A. Transcription factories: gene expression in unions? Nat. Rev. Genet. 10, 457–466 (2009).
Melnik, S. et al. The proteomes of transcription factories containing polymerases I, II, or III. Nat. Methods 28, 963–968 (2011).
Caudron-Herger, M., Cook, P.R., Rippe, K. & Papantonis, A. Dissecting the nascent human transcriptome by analyzing the RNA content of transcription factories. Nucleic Acids Res. 43, e95 (2015).
Jackson, D.A., Iborra, F.J., Manders, E.M.M. & Cook, P.R. Numbers and organization of RNA polymerases, nascent transcripts and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 (1998).
Kimura, H., Tao, Y., Roeder, R.G. & Cook, P.R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 19, 5383–5392 (1999).
Jackson, D.A. & Cook, P.R. Transcription occurs at a nucleoskeleton. EMBO J. 4, 919–925 (1985).
Ahmad, Y., Boisvert, F.M., Gregor, P., Cobley, A. & Lamond, A.I. NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res. 37, D181–D184 (2009).
Aviner, R., Geiger, T. & Elroy-Stein, O. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 27, 1834–1844 (2013).
Core, L.J., Waterfall, J.J. & Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Nojima, T. et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Bhatt, D.M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012).
Ameur, A. et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 18, 1435–1440 (2011).
Khodor, Y.L. et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 25, 2502–2512 (2011).
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 (2011).
Windhager, L. et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 22, 2031–2042 (2012).
Kim, T.K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Kaikkonen, M.U. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013).
Mann, M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 (2006).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Besse, S., Vigneron, M., Pichard, E. & Puvion-Dutilleul, F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 4, 143–161 (1995).
Novakova, Z., Man, P., Novak, P., Hozak, P. & Hodny, Z. Separation of nuclear protein complexes by blue native polyacrylamide gel electrophoresis. Electrophoresis 2, 1277–1287 (2006).
Nadano, D., Aoki, C., Yoshinaka, T., Irie, S. & Sato, T.A. Electrophoretic characterization of ribosomal subunits and proteins in apoptosis: specific downregulation of S11 in staurosporine-treated human breast carcinoma cells. Biochemistry 40, 15184–15193 (2001).
Raposo, R.A.S., Thomas, B., Ridlova, G. & James, W. Proteomic-based identification of CD4-interacting proteins in human primary macrophages. PLoS ONE 6, e18690 (2011).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Trudgian, D.C. et al. CPFP: a central proteomics facilities pipeline. Bioinformatics 26, 1131–1132 (2010).
Shteynberg, D. et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690 (2011).
Lovén, J. et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012).
Madsen, J.G. et al. iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 43, e40 (2015).
Gaidatzis, D., Burger, L., Florescu, M. & Stadler, M.B. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 33, 722–729 (2015).
Acknowledgements
We thank B. Deng and J. Bartlett for their help, and the Sir William Dunn School of Pathology (SWDSOP) proteomics and BioQuant/DKFZ sequencing facilities for the high-throughput analyses. This work was supported by a Medical Research Council grant (to P.R.C.), by the ERASysBio+/FP7 initiative (to K.R. and P.R.C.), by a DKFZ intramural grant (to M.C.-H.) and by Center for Molecular Medicine core funding (to A.P.).
Author information
Authors and Affiliations
Contributions
S.M. and P.R.C. conceived and developed the factory isolation procedure. S.M., A.P., I.M.C. and P.R.C. implemented and validated the procedure. M.C.-H., L.B., K.R. and A.P. adapted and implemented the protocol for nascent RNA isolation. All authors analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Melnik, S., Caudron-Herger, M., Brant, L. et al. Isolation of the protein and RNA content of active sites of transcription from mammalian cells. Nat Protoc 11, 553–565 (2016). https://doi.org/10.1038/nprot.2016.032
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.032
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.